Protein Tagging: How to Choose a Tag for Your Protein
Sara Klink
Promega Corporation
Publication Date: Revised 4/23; tpub_215
Abstract
Protein tagging is an important aspect of protein analysis. Being able to tag your protein for various applications, including purification, pull-down assays, immunoassays and more can help you define protein function. But which tag should you choose? This article covers several options for affinity-, epitope- or reporter-based tags and reagents that can help you with your experiments.
Introduction
Proteins or polypeptides are integral to cellular function. They are the enzymes that make up pathways for energy metabolism, cellular growth, signaling, mitotic cycle and cell death. Being able to isolate and detect specific proteins allows us to understand which proteins are part of larger complexes, which proteins interact and their role in cellular function and disease means. One way to examine the function of a protein is by adding a tag to the protein of interest and separating it from a cell lysate to study the protein.
Fusion tags can be polypeptides, small proteins or enzymes added to the amino (N) or carboxy (C) terminus of a protein. Tagging can be done via cloning into vectors or added using CRISPR-Cas9 gene editing to tag an endogenous protein. By using an affinity tag, you can isolate or immobilize a protein for additional proteomic studies.
While researchers commonly tag a protein to purify it from a cellular lysate and use the isolated protein in biochemical assays, a peptide tag can do more. For example, epitope-tagged proteins can be detected with a tag-specific antibody if there are no antibodies specific to your protein. In addition, tags can also quantitate proteins, study protein complexes and determine if the protein is inside or outside the cell. You can even visualize protein location within a cell because of the tag on your protein.
So which protein tag is right for your experiments? Below is a list of options for tagging proteins with tools that can help.
Polyhistidine
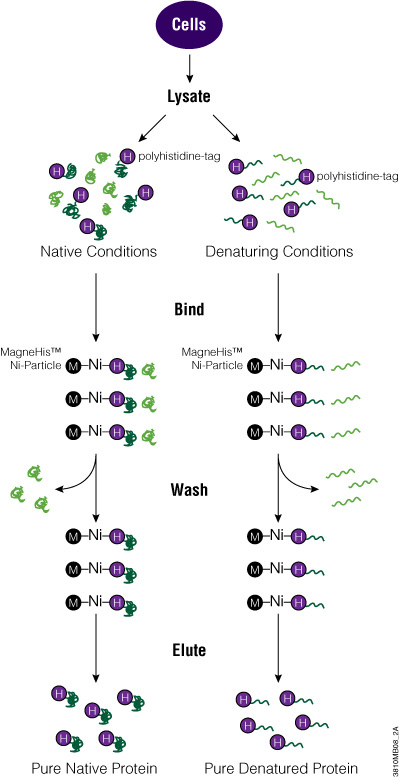
One of the simplest and smallest tags is the polyhistidine tag or 6XHis tag. Typically, this peptide tag is made up of six histidine amino acids that can be placed on the N or C terminus of a protein. Because of its small size, the His tag tends not to alter the structure of a protein, which is useful for downstream assays. In addition, the His tag can bind to metal ions like Ni2+, Zn2+ and Cu2+, making it useful for purifying or immobilizing fusion proteins.
The polyhistidine tag works well to rapidly purify proteins expressed in E. coli, but for proteins expressed in mammalian or insect cells, the greater number of histidine residues means there is higher background binding. Therefore, to increase protein purity, the metal resins that bind His-tagged proteins need to be stringently washed. In addition, denaturing agents like 8M urea or 6M guanidine HCl can be used with His-tagged membrane or insoluble proteins to improve purification.
To add the His tag to your protein, clone the ORF into a vector that carries the tag. Depending on the promoter used, express the tagged protein in bacterial, mammalian or insect cells. Alternatively, you can use cell-free expression systems for protein expression. To purify the protein, you can use magnetic beads (e.g., MagneHis™ Protein Purification System) or nonmagnetic resins (e.g., HisLink™ Protein Purification System) for cell lysates. Once eluted from the purification matrix, your tagged protein is ready to be used for further investigation.
Polyhistidine-Tagged Protein Purification
Glutathione-S-Transferase
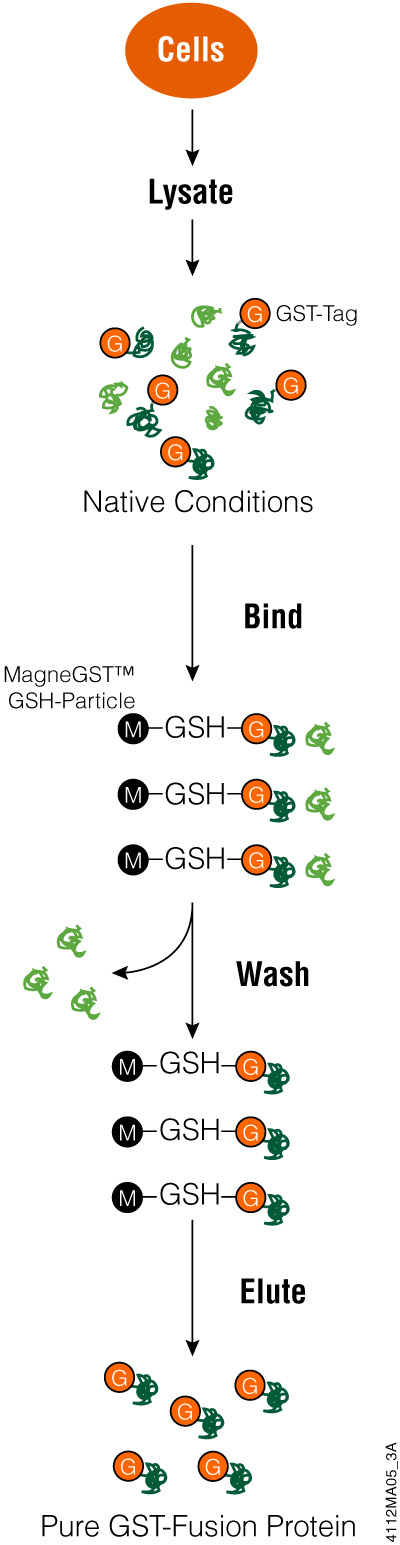
Glutathione-S-Transferase (GST) is an affinity tag used for proteins expressed in E. coli to purify the tagged proteins from bacterial lysates. This 26kDa peptide tag is part of a family of cystolic proteins present in eukaryotes, making it less useful when attempting to isolate proteins from eukaryotic cells because of competition from other proteins. Tagging eukaryotic proteins with GST enhances fusion protein solubility when expressed in bacteria. In addition, GST-tagged proteins can be expressed at high levels in bacteria but may form inclusion bodies due to protein aggregation. The GST tag can be added to the N or C terminus of the protein of interest.
GST has a strong affinity for glutathione, meaning you can capture GST protein fusions on an immobilized matrix like glutathione-coated beads. This binding characteristic is used for protein purification as well as capturing proteins that might bind to your protein (e.g., pull-down assays).
While proteins with a GST tag can be highly expressed and more soluble, the large size of the tag can interfere with protein function in downstream applications. You may be able to cleave your protein from the GST tag, reducing possible functional interference. If the expressed protein has aggregated into an inclusion body, affinity purification using glutathione becomes problematic. GST needs to be correctly folded for successful binding to the glutathione and even if the GST tag refolds, your protein of interest may not be.
To use the GST tag, start by cloning the coding region for the protein of interest into a vector and express the tagged protein in E. coli. Using an inducible promoter to control target protein expression can help with proteins that are toxic to bacterial cells. Once bacterial cells are lysed (e.g., using FastBreak™ Cell Lysis Reagent), you can purify the GST-tagged protein using a glutathione-based resin. Depending on the cloning vector used, there may be a cleavage site for a protease to remove the GST tag from your protein. Once your protein is eluted or cleaved, it is ready for analysis.
Purifying GST-Protein Fusions
HaloTag® Protein
While GST and 6XHis tags are useful for isolating proteins expressed in bacterial cells, there are tagging options that are compatible with both prokaryotes (e.g., E. coli) and eukaroytes (e.g., mammalian cells). The 34kDa HaloTag® protein offers this expression system flexibility. In contrast to other tags that use noncovalent interactions to purify proteins, the basis of HaloTag® technology is the covalent binding between HaloTag® protein and its ligand. Because of the strength of the covalent bond, you can wash your tagged protein under stringent conditions and basically eliminate any nonspecific protein background. Worried about the size of the tag affecting the function of your protein? No problem! Just use TEV protease to cleave your protein from HaloTag and the purified, untagged protein is ready to use in downstream analysis. Learn more about HaloTag® Technology here.
One of the challenges of expressing proteins in bacterial cells is solubility. Eukaryotic proteins are not methylated and lack other post-translational modifications when synthesized in E. coli, making them prone to forming inclusion bodies. Fusion tags like that of HaloTag® protein can make recombinant proteins more soluble and even enhance expression in bacteria, making your protein of interest easier to purify. By expressing your HaloTag®-protein fusion in mammalian cells, post-translational modifications will be present, more closely reflecting how the protein will function under cellular conditions. Using an immobilized matrix like resin coated in HaloTag® ligand, you can purify only proteins with HaloTag present or use in pull-down assays for discovering what protein or proteins bind to your protein of interest. In this way, you can better understand protein interactions.
But the applications don’t stop with protein:protein interactions and protein purification. If you want to use immunochemistry, we have an antibody for that. The Anti-HaloTag® pAb is a good place to start.
Interested in cell imaging? You will need to pick the right fluorescent ligand (e.g., HaloTag® Ligands or Janelia Fluor® HaloTag® Ligands). With your protein fused to HaloTag® protein and fluorescent HaloTag® ligands, the fluorescent dyes provide labeling or detection options for studying protein:protein interactions, protein localization, protein trafficking and turnover, and cell counting using FACS. The brightness and stability of the Janelia Fluor® HaloTag® Ligands means they are especially useful for intracellular imaging, including single-molecule imaging and tracking in live or fixed cells, confocal fluorescent imaging and super resolution microscopy techniques including dSTORM.
Not seeing the ligand you need for your project? Create virtually any HaloTag® Ligand you can imagine using the HaloTag® Ligand Building Blocks. The chloroalkane group to which HaloTag® protein covalently bonds can be applied to any compound or surface with a compatible chemical group.
To get started with HaloTag® technology, clone the coding region for your protein of interest in traditional or Flexi® cloning vectors to add HaloTag at the N or C terminus. Alternatively, you can search for any of the thousands of prebuilt HaloTag® clones available from Kazusa. Once you have your clone, you can use it for any protein analysis application. Depending on what you want to do next, you may need a purification system (e.g., HaloTag® Mammalian Protein Purification System or HaloTag® Protein Purification System) or a pull-down assay (e.g., HaloTag® Mammalian Pull-Down Systems). There are bright fluorescent HaloTag® Ligands or Janelia Fluor® HaloTag® Ligands to use for imaging studies, the Anti-HaloTag® pAb for immunological detection and the NanoBRET™ PPI Systems to examine protein:protein interactions.
HaloTag® Interchangeable Labeling Technology

While most of the peptide tags discussed previously are affinity tags for use in protein purification, pull-down assays or methods that bind the tagged protein to a solid matrix, there are other types of tags for different applications. From quantitating proteins to endogenous tagging, a bioluminescent tag or even a small reporter protein fused to a protein can be used to track a tagged protein in the cell, eliminating the need for antibodies.
HiBiT Peptide Tag
Smaller is better for many things and a tiny 11 amino acid peptide tag called HiBiT is great for tagging a protein. If you want to skip traditional cloning, you can even use CRISPR gene editing to add the HiBiT tag to an endogeneous protein. This has the advantage of studying a protein without overexpression typically seen when using exogenous expression from a plasmid. By keeping the cellular proteins in the same proportions found in the cell, you are able to study changes in your tagged protein under physiological conditions. This advantage means fewer artifacts from protein overexpression and more biologically relevant results. The CRISPR protocol for adding the peptide tag is simple and the small size if HiBiT offers high editing efficiency.
On its own, HiBiT is not bioluminescent. However, once a detection reagent that contains the complementary LgBiT protein is added, a bright luminescent signal is produced. The quantifiable signal is easy to measure using a luminometer. Bioluminescent assays are sensitive and offer detection down to nanomolar levels. Want to examine extracellular or secreted proteins? We have a reagent system for that! Combine quantitating total labeled protein with our Lytic System and examine secreted protein levels using our Extracellular system. You can even eliminate the need for an antibody and use our bioluminescent blotting system to detect proteins blotted to a membrane. If LgBiT is expressed intracellularly, you can kinetically detect HiBiT-tagged protein abundance in live cells. See how you can use HiBiT for protein tagging here.
Overview of the Nano-Glo® HiBiT Lytic Detection System

HiBiT also makes an excellent epitope tag for those who would like to pursue more traditional immunological assays, expanding the protein analysis options beyond bioluminescent assays. For endogenous HiBiT-tagged proteins, the Anti-HiBiT Monoclonal Antibody (mAb) can confirm subcellular localization. Perform Western blotting to assess protein levels and size for proteins tagged with HiBiT. Immunoprecipitate HiBiT-tagged proteins to enrich for protein of interest or determine binding partners. You can even use the HiBiT tag and antibody for fluorescence-activated cell sorting (FACS) analysis on live or fixed cells. With subpicogram sensitivity and high specificity, the Anti-HiBiT mAb performs as well or better as other commonly used epitope tag antibodies, adding to the versatility of the HiBiT tag.
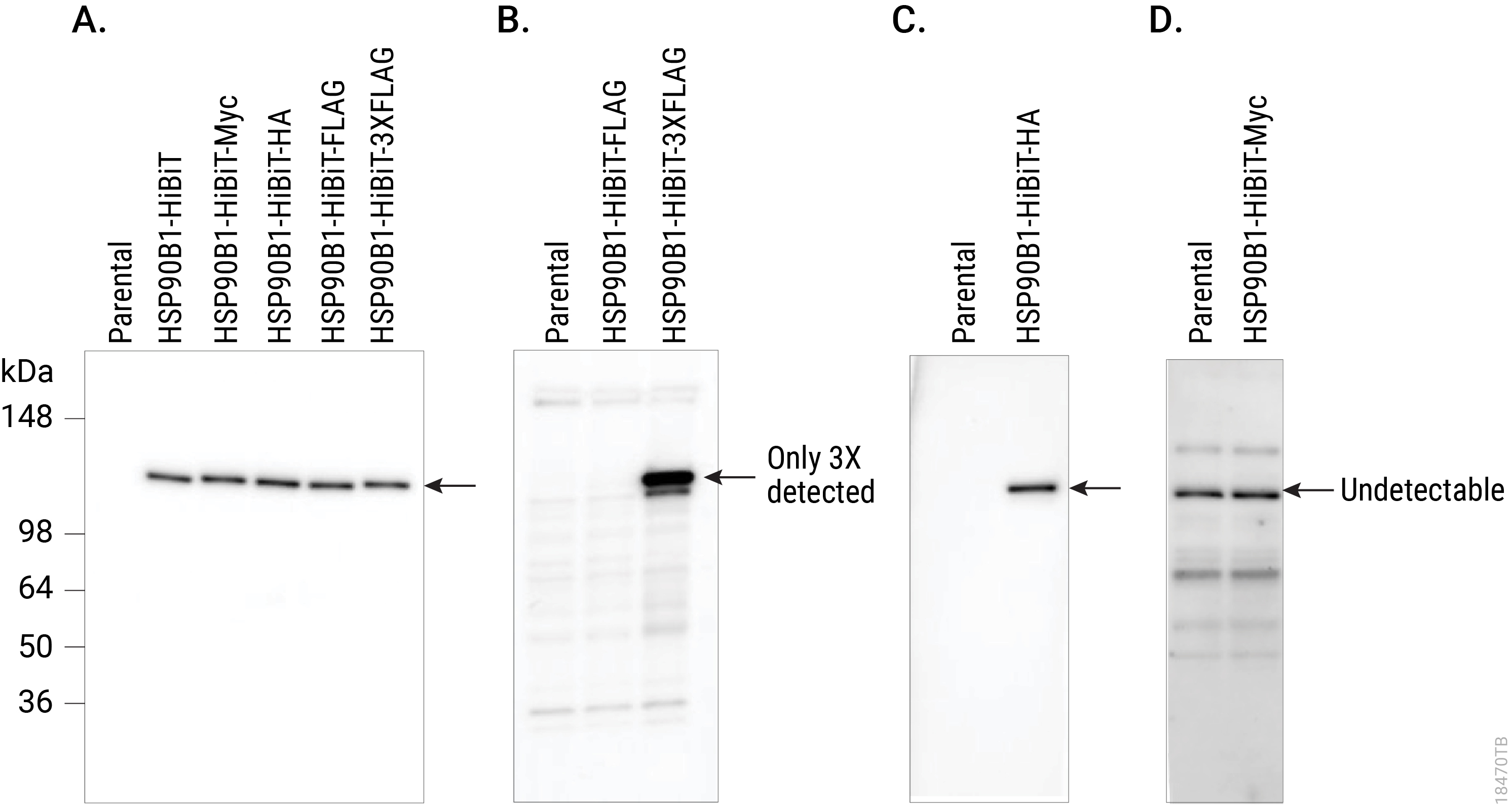
Comparing Western blot detection using Anti-HiBiT Monoclonal Antibody to common epitope tag antibodies. HSP90B1 was endogenously tagged on the C terminus in HeLa cells with HiBiT alone or HiBiT and a single copy of the Myc, HA or FLAG® tags, or three copies of the FLAG® tag. Lysates (10µg) from the HSP90B1-tagged cell pools were separated by SDS-PAGE. After transfer to PVDF membrane, Western blotting was performed with 1µg/ml Anti-HiBiT Monoclonal Antibody (Panel A), 10µg/ml Anti-FLAG® (M2; Panel B), 0.1µg/ml Anti-HA (3F10; Panel C) or 1µg/ml Anti-Myc (9E10; Panel D) primary antibodies and 0.4µg/ml anti-mouse or anti-rat (for anti-HA blot) secondary antibody and ECL substrate using a 5-minute exposure. The HiBiT antibody image brightness was scaled 0–29,817 and all other images 145–2,735.
To start using HiBiT, you can clone your ORF into one of our traditional MCS or Flexi® vectors or use a CRISPR method to add the tag to the genomic sequence. Place the tag where needed for optimal detection. Once your protein is tagged with HiBiT, determine which method works best for your studies (Nano-Glo® HiBiT Lytic, Extracellular or Blotting Systems). Alternatively, detect HiBiT using a sensitive antibody-based method with the Anti-HiBiT Monoclonal Antibody.
NanoLuc® Luciferase
Sometimes only a reporter will do. Luckily, NanoLuc® luciferase is a 19kDa enzyme, making the bioluminescent protein small enough to hitch a ride with another protein without interfering with normal cellular functions. Similar to HiBiT, NanoLuc provides a sensitive, easily quantified protein fusion tag. However, the reporter enzyme does not require a complementation partner, simplifying its use in some applications. The brightness and small size means NanoLuc® luciferase can fit into viral particles without causing difficulties. Read how these virology researchers did it. As a reporter enzyme, NanoLuc® luciferase can help detect the cellular location of a fusion protein as well as tracking the path your protein might take. In addition, NanoLuc® fusion proteins can be used in bioluminescence resonance energy transfer (BRET)-based applications for studying protein:protein interactions or small molecule target engagement. Learn more about the capabilities of this reporter enzyme. Alternatively, use the Anti-NanoLuc® Monoclonal Antibody for detecting NanoLuc® luciferase in Western blotting and immunofluorescence.
To create a protein fusion with NanoLuc® luciferase, you will need to clone your ORF into a NanoLuc® reporter vector, placing NanoLuc on the N or C terminus. Depending on your application, you could use a lytic detection system (e.g., Nano-Glo® Luciferase Assay System) or a live-cell detection assay (e.g., Nano-Glo® Live Cell Assay System).
Summary
Choosing a tag for your protein is based on your experimental needs. Whether you need an affinity tag for protein purification, want to create a single fusion protein to be used in a variety of protein analysis methods, or add an endogenous tag using CRISPR-Cas9 gene editing, there are an array of tags and reporter enzymes available.
Additional Protein Tagging Resources

An Overview of Affinity Tags
This article describes various affinity tags and covers their use for protein purification.
Tools for Protein Analysis
This proteomics guide covers products for investigating protein function.