RAS Pathway Drug Discovery
RAS is the most frequently mutated oncogene in human cancers, and KRAS is the most frequently mutated subtype of the RAS family. Most lung, pancreatic and colorectal cancers are driven by KRAS mutations. Until the recent discovery of KRAS (G12C)-specific covalent inhibitors, KRAS was considered an undruggable target.
Promega offers several assays and technologies to facilitate RAS drug discovery, including assay collections for the interrogation of and profiling against hot-spot mutants. These assays measure the binding of compounds to pathway components and assess their impact on protein:protein interactions, protein levels and, ultimately, activation of ERK.
Interested in more information about RAS pathway drug discovery?

Learn how Target Engagement Assays are used to study the RAS pathway in this poster: Comprehensive TE assays for the RAS pathway and its proximal effectors including SHP2
Active, GTP-bound Ras activates RAF, followed by phosphorylation and activation of MEK, which is followed by phosphorylation and activation of ERK.
- RAS: A family of small GTPases that includes KRAS, HRAS and NRAS
- RAF: A serine/threonine kinase family consisting of c-Raf, B-Raf and A-Raf
- MEK: A mitogen activated protein kinase kinase (MAPK/ERK Kinase)
- ERK: An extracellular signal-regulated kinase family, also known as a mitogen activated protein kinase (MAPK), consisting of ERK1 and ERK2 that share ~85% sequence identity

An overview of the RAS-RAF-MEK-ERK pathway.
Target Engagement
NanoBRET® technology offers a sensitive, specific method to measure the interaction of small-molecule drugs with their target protein in live cells.
With the NanoBRET® Target Engagement Assay, you can:
- Quantitate compound affinity (how tightly it binds to a protein) and target protein occupancy (how much compound binds to a protein) in live cells.
- Assess how long a compound binds to the target protein (its residence time) under physiological conditions.
- Scale the simple, multiwell assay to suit your research throughput needs.
- Generate high‐quality data with low error rates and high reproducibility.
- Get started quickly with available ready-to-use assays.

Principle of the NanoBRET® Target Engagement Assay.
Learn more about NanoBRET® Target Engagement
Measure TE on Intracellular RAS Complexes
The NanoBRET® TE Intracellular RAS Assay measures intracellular compound binding to RAS proteins with ready-to-use assays for quantify compound affinity and fractional occupancy for multiple RAS variants, including KRAS, HRAS and related hot-spot mutants. Utilizing NanoLuc® Binary Technology (NanoBiT) complementation to create the energy donor enables the measurement of cellular target engagement on multimeric RAS complexes to more accurately reflect RAS biology. Measure engagement for individual RAS complexes or profile compound selectivity across RAS variants.


Learn more about our NanoBRET® TE Intracellular RAS Assays
Featured Publication
In this article, published in Nature Chemical Biology, we investigated the reversible binding of KRAS small-molecule inhibitors to assess which hotspot mutants are vulnerable to switch-II pocket (SII-P) engagement in cells. We showed that SII-Ps of many KRAS hotspot mutants are accessible using noncovalent ligands. Our results emphasize the SII-P as a privileged drug-binding site on KRAS and reveal new therapeutic opportunities in RAS-driven cancer.

Conditionally Quantify TE at RAF PPI Dimers
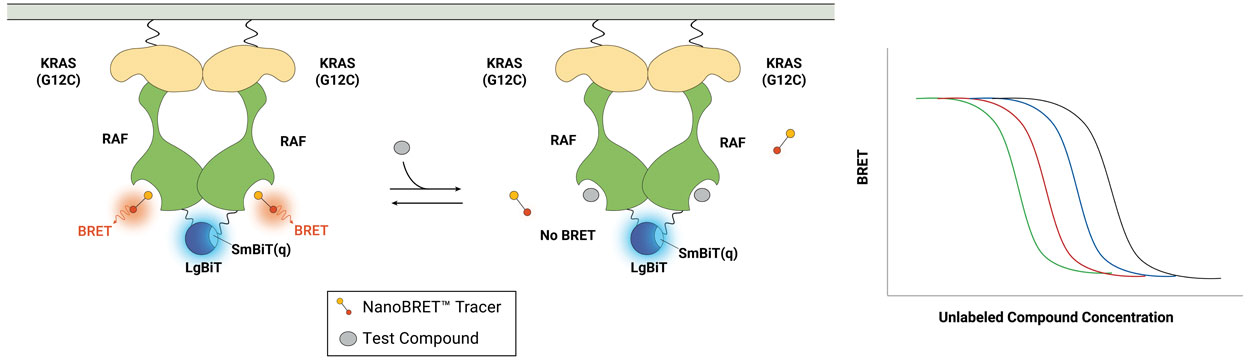

Featured Publication
RAF dimer inhibitors show promise in treating RAS-driven cancers by targeting RAF protomers in the RAS-RAF complex. In this paper, learn how a new method quantifies drug occupancy in active RAS-RAF complexes, aiding inhibitor assessment. Clinical-type II RAF inhibitors primarily engage the ARAF protomer, impacting MAPK signaling in mutant RAS cell lines, revealing ARAF's critical role.

For a complete list of Kinase Target Engagement Assays including RAF, MEK and ERK, see our Selection Table.
Degradation
By using CRISPR/Cas9 gene editing to knock in a bioluminescent tag at endogenous loci of interest, proteins expressed by the native promoter can be studied using simple light output measurements. Compared to overexpression models, endogenously tagged proteins maintain natural epigenetic regulation and stoichiometry with native interacting partners, resulting in improved assay windows, minimized artifacts, and more accurate models of protein biology under physiologically relevant conditions.

An illustration of HiBiT:LgBiT complementation to study protein degradation within a cell.
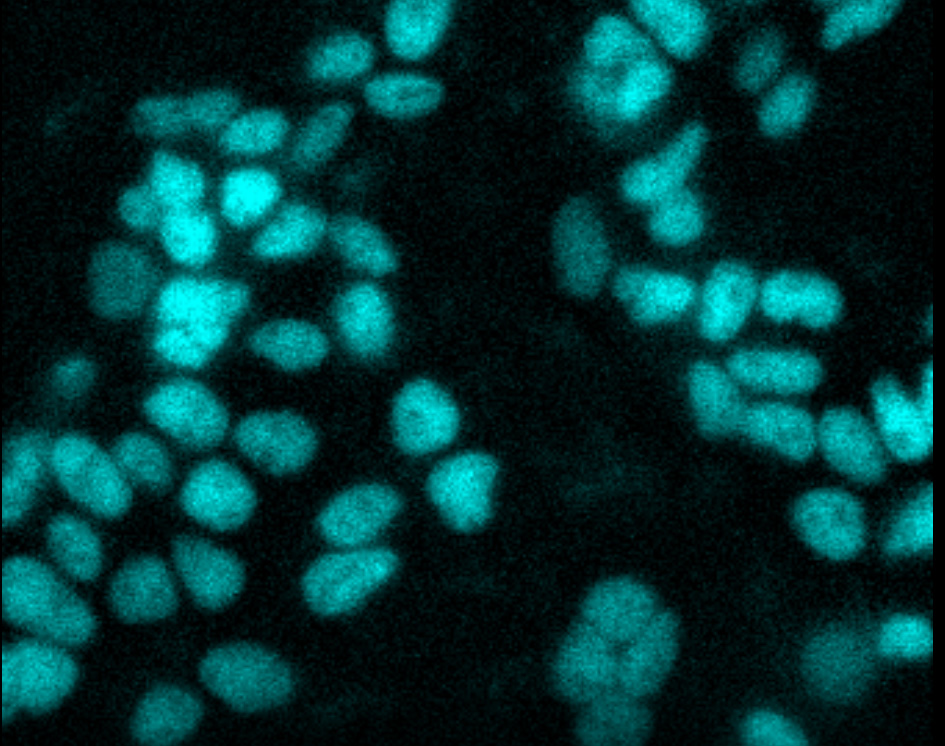
Time-lapse live-cell imaging. CRISPR-HiBiT BRD4 cells were treated with MZ1, a BET bromodomain degrader. Uniform loss of BRD4 was observed over 2 hours. Imaging was performed using an Olympus LV200 System.
HiBiT Knock-in Cell Lines for RAS Pathway Targets
HiBiT is a popular tag for studying endogenous protein degradation. HiBiT is a small, 11 amino acid peptide that binds with high affinity to a larger subunit called LgBiT. The bound complex has high luciferase activity and will generate a bright luminescent signal in the presence of added substrate. The small size of the HiBiT tag makes it well suited for endogenous tagging via CRISPR/Cas9 editing.
Addition of compounds that elicit degradation of the tagged protein results in loss of luminescence signal, which is highly quantitative and can be measured in real time under conditions with intracellular LgBiT. Cellular dose-response curves can be obtained and monitored over a 24- to 48-hour time frame, allowing for accurate determination of degradation rate, Dmax, DC50 values and protein recovery. This approach allows rapid rank ordering of degradation against a series of different parameters, and the assay is suitable for high-throughput screening.
A.

B.

Live cell degradation kinetics of endogenous HiBiT-KRasG12C following PROTAC treatment. Panel A. HiBiT was inserted via CRISPR/Cas9 at the N-terminal endogenous KRasG12C loci in the MIA-PaCa2 cell line. Following LgBiT expression, dose-response kinetic degradation experiments were performed using the VHL-based KRasG12C PROTAC, LC-2, in CO2-independent medium containing Nano-Glo® Endurazine™ Substrate. Fractional RLU is plotted relative to the DMSO control. Panel B. Similar live-cell luminescent studies of HiBiT-KRasG12C in MIA-Paca2 cells using the parent inhibitor, MRTX849, which does not show loss of KRasG12C over the same dose-response treatments.
Need to quantify protein levels in disease-relevant cellular models?
We provide ready-to-use HiBiT knock-in cell lines and pools, including WT and mutants for KRAS, NRAS, ARAF, BRAF and RAF1.| KI Cell Line | Background |
|---|---|
| HiBiT-KRAS(G12C) | MIA PaCa-2 |
| HiBiT-KRAS(G12D) | AsPc-1 |
| HiBiT-KRAS(G12D) | SW1990 |
| HiBiT-KRAS(G12D) | HEK293 |
| HiBiT-KRAS(G12S) | A549 |
| HiBiT-KRAS(G12V) | HEK293 |
| HiBiT-KRAS(G12V) | SW620 |
| HiBiT-KRAS(G13D) | NCI-H647 |
| HiBiT-KRAS(WT) | HEK293 |
| HiBiT-KRAS(WT) | NCl-H1299 |
| HiBiT-NRAS(Q61R) | SW1271 |
| HiBiT-NRAS(WT) | HEK293 |
| HiBiT-ARAF | A549 |
| HiBiT-BRAF | A549 |
| HiBiT-BRAF(V600E) | A375 |
| RAF1-HiBiT | A549 |
| RAF1-HiBiT | Hs766T |
| RAF1-HiBiT | PANC 08.13 |
Protein Interactions
NanoBRET™ Technology
NanoBRET™ technology enables sensitive, reproducible detection of protein-protein interactions (PPI) in the natural cellular environment. The use of full-length proteins expressed at low levels enables PPI monitoring and screening studies that reflect true cellular physiology. The bright, blue-shifted donor signal and red-shifted acceptor create optimal spectral overlap, increased signal and lower background compared to conventional BRET assays.

Principle of the NanoBRET™ PPI assay.
You can read more about the capabilities of NanoBRET™ technology and see example data in this publication:
Machleidt, T. et al. (2015) NanoBRET—A novel BRET platform for the analysis of protein–protein interactions. ACS Chem. Biol. 10(8), 1797–1804.
See how NanoBRET™ PPI is used to study the RAS pathway in this poster.
A.

B.

NanoBRET™ assays across RAS and RAF family members for the study of inhibition and/or induction of protein interactions in the pathway. Kinetic RAS:RAF assays +/– BI-2852 inhibitor (Panel A); RAF dimerization assays showing induction with GDC0879 (Panel B).
A.

B.

Inhibition of KRAS WT:SOS1cat domain interaction. SOS1 catalytic domain shows dissociation from KRAS WT with dose response treatment of BI-3406 and BAY-293.
NanoLuc® Binary Technology (NanoBiT) is a two-subunit system based on NanoLuc® luciferase that can be applied to the intracellular detection of PPIs in live cells. This short video explains how the technology works.
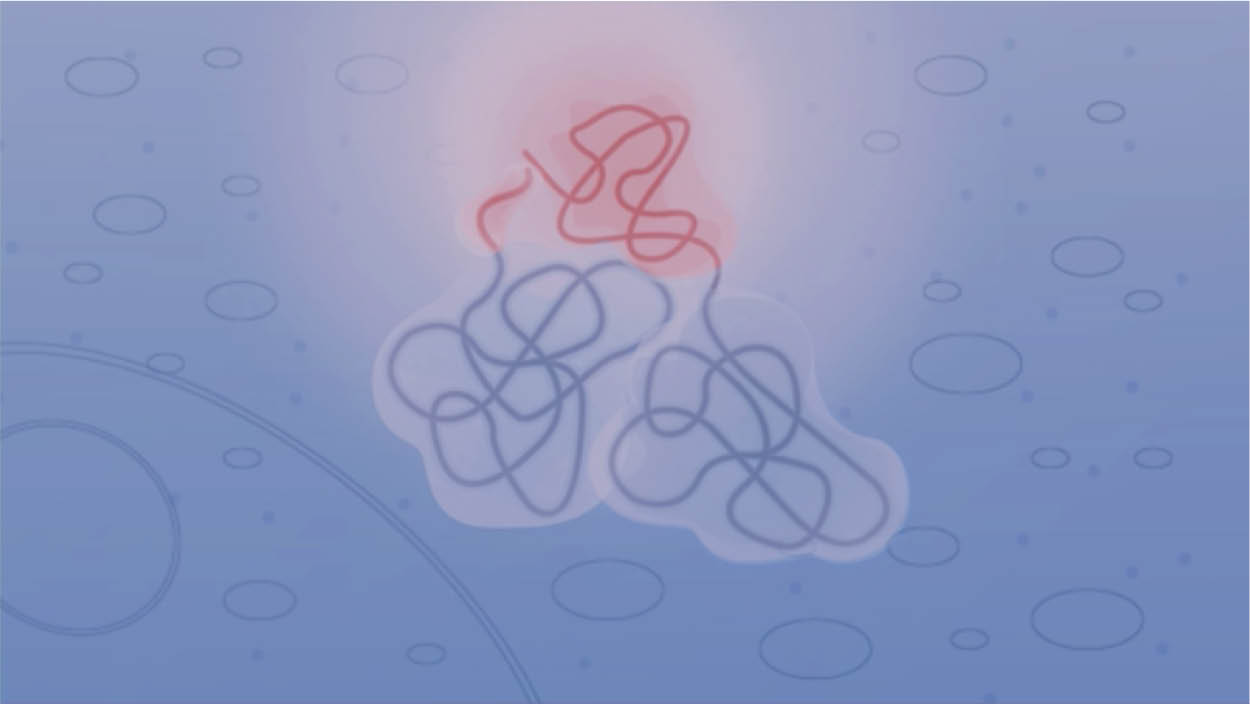

Principle of the NanoBiT® PPI assay.
To learn more about using NanoBiT® technology to study PPIs involved in the RAS pathway, download this slide deck showing data from wild-type and mutant KRAS paired with CRAF or BRAF.
Read about the origins of NanoBiT® technology in this paper: Dixon, A. S. et al. (2016). NanoLuc Complementation Reporter Optimized for Accurate Measurement of Protein Interactions in Cells. ACS Chemical Biology. p. 400-408



NanoBiT® assays to monitor interaction of KRAS 4B wild-type and mutants with RAF isoforms. KRAS 4B with the CRAF RBD domain (Panel A); KRAS 4B with full-length CRAF (Panel B); KRAS 4B with full-length BRAF (Panel C).

Real-time monitoring of KRAS 4B (WT):CRAF (full-length) interaction in serum-starved HeLa cells following EGF treatment.
We have a variety of NanoLuc®, HaloTag®, NanoBiT® and BiBRET vectors to help you get started with your PPI assays. See all available assays for interrogating RAS pathway interactions in live cells.
Detect PPI of Tagged Proteins

In the Lumit® Anti-Tag Protein Interaction Reagents, streptavidin and antibodies against common tags are labeled with LgBiT and SmBiT. When two proteins with different tags bind, the interaction can be detected using the corresponding SmBiT- and LgBiT-conjugated antibodies.


KRAS(G12C)/RBD-cRAF Interaction with AMG510

Lumit® KRAS-cRAF assay can be used to test the specificity of small molecule inhibitors for panels of KRAS mutants.
Kinetics of KRAS(G12C)/RBD-cRAF interaction

The KRAS activation cycle can be reconstituted in vitro and the kinetics of KRAS/cRAF binding can be monitored in the presence of small molecule inhibitors as a reflection of SOS1-mediated GDP/GTP exchange.
To see more data, view the application note: Modulating KRAS-c-RAF Interaction Using Small Molecule Inhibitors or visit the Lumit® Anti-Tag Protein Interaction Reagents product page.
Phospho-ERK
The Lumit® Immunoassay Cellular System is a no-wash bioluminescent immunoassay that measures target analytes directly in cell lysates. No media removal or lysate transfer—just add labeled antibodies to the sample, add the detection reagent and read the luminescent signal—all in a single plate! We offer a complete assay for detecting phospho-ERK activity that includes all reagents and antibodies.

The principle of the Lumit® Immunoassay Cellular System.

You can learn more about using the Lumit® p-ERK Immunoassay to study KRAS in this publication:
Swiatnicki, M. et al. (2022) Profiling oncogenic KRAS mutant drugs with a cell-based Lumit p-ERK immunoassay. SLAS Discovery. 27(4), 249–57.

RAS mutant inhibitors. Lumit® assay demonstrates cellular p-ERK inhibition profile of individual mutant KRAS inhibitors. Titrations of various mutant KRAS inhibitors were completed in MCF-7 (WT), SW620 (G12V mutant), Mia-PaCa-2 (G12C mutant), AsPC-1 (G12D mutant), with Lumit® p-ERK being used as a proxy to detect KRAS inhibition. Panels A–C. High selectivity of KRAS G12C compounds is shown through p-ERK Lumit® assay, with complete inhibition of Mia-PaCa-2 cells and no inhibition of KRAS WT, G12D or G12V cells. Panel D. Lumit® assay shows MRTX-1133 is very selective for KRAS G12D with some promiscuity towards KRAS G12C and G12V at higher IC50s.
Learn about our Lumit® Immunoassay Cellular Systems—Complete Assay for detecting phospho-ERK that includes all reagents, including antibodies.
GTPase
The GTPase-Glo™ Assay analyzes the intrinsic activities of GTPase, GAP-stimulated GTPase, GAP and GEF. The assay measures enzymatic conversion of GTP remaining, after the GTPase reaction, into ATP followed by bioluminescent detection of the created ATP.
To see more data, read this publication, or visit the GTPase-Glo™ Assay product page.
Profiling Services
Our custom assay services can help to accelerate your drug discovery and development workflow. These comprehensive services are built around our technologies for sensitive, high-throughput and biologically relevant results. For more information, contact the Tailored R&D Solutions team.
- Live cell target engagement
- CRISPR-HiBiT cell lines
- Protein-protein interactions
- Lumit® immunoassays
- And more