Kinase Biology for Drug Discovery
Protein and lipid kinases are involved in a wide variety of pathological conditions. Accordingly, kinase biology constitutes a critical field for drug discovery research. Several bioluminescent assays are available to query the ability of compounds to bind or inhibit kinases in biochemical and cellular formats, amenable to high throughput or selectivity profiling.
Need help with cellular kinase profiling? Learn about our services.
Kinase Biochemical Activity Assays
Schematic Overview of the ADP-Glo™ Kinase Assay
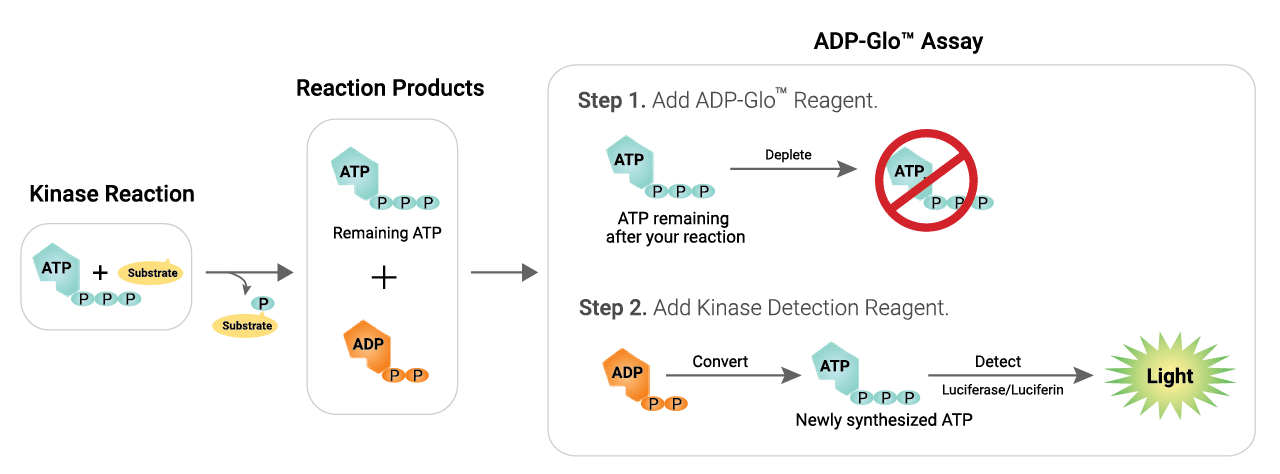
Detect Activity of a Range of Kinases
The ADP-Glo™ Kinase Assay has a high dynamic range and produces a strong signal at low ATP to ADP conversion, making it well suited for screening low activity kinases such as growth factor receptor tyrosine kinases. The assay can be used at ATP concentrations up to 1mM, important for kinases with high Km values for ATP.
The ADP-Glo™ Assay is a universal assay that works with any kinase and substrate. Representative examples of kinase activity curves are shown here.




View our complete range of kinase biochemical assays.
Kinase Enzyme Systems
Easily screen and profile kinase inhibitors with Kinase Enzyme Systems.
- Currently 379 kinases available, with 100+ mutants!
- Each Kinase Enzyme System includes pre-qualified enzyme, substrate and reaction buffer optimized for use with ADP-Glo™ Assays
- All Kinase Enzyme Systems have Application Notes available so you can get started quickly
- Kinases are available in strip format for easier profiling (see Kinase Selectivity Profiling Systems)
The Kinase Enzyme Systems Span the Breadth of the Human Kinome

View all Kinase Enzyme Systems
NanoBRET® TE Kinase Assays
The NanoBRET® Target Engagement (TE) Intracellular Kinase Assay is based on the NanoBRET® System, an energy transfer technique designed to measure molecular proximity in living cells. The NanoBRET® TE Assay measures the apparent affinity of test compounds by competitive displacement of the NanoBRET® tracer, reversibly bound to a NanoLuc® luciferase-kinase fusions in cells.
- More than 340 assays available.
- Quantitatively measure test compound affinity and fractional occupancy for kinases in live cells for more physiologically relevant information.
- These assays measure the interaction between full-length kinases and test compounds.
- Ready-to-use, kinase-specific assays offer a simple workflow and high-throughput compatibility
- Detect multiple kinase inhibitor types, such as inhibitors with different binding modes (including type I, II, allosteric and covalent)
- Measure the durability of test compound binding to kinase (residence time) using kinetic analysis

Applications of NanoBRET® TE Kinase Assays. The assays can measure test compound affinity under equilibrium conditions (upper panel), or residence time under non-equilibrium conditions (lower panel).
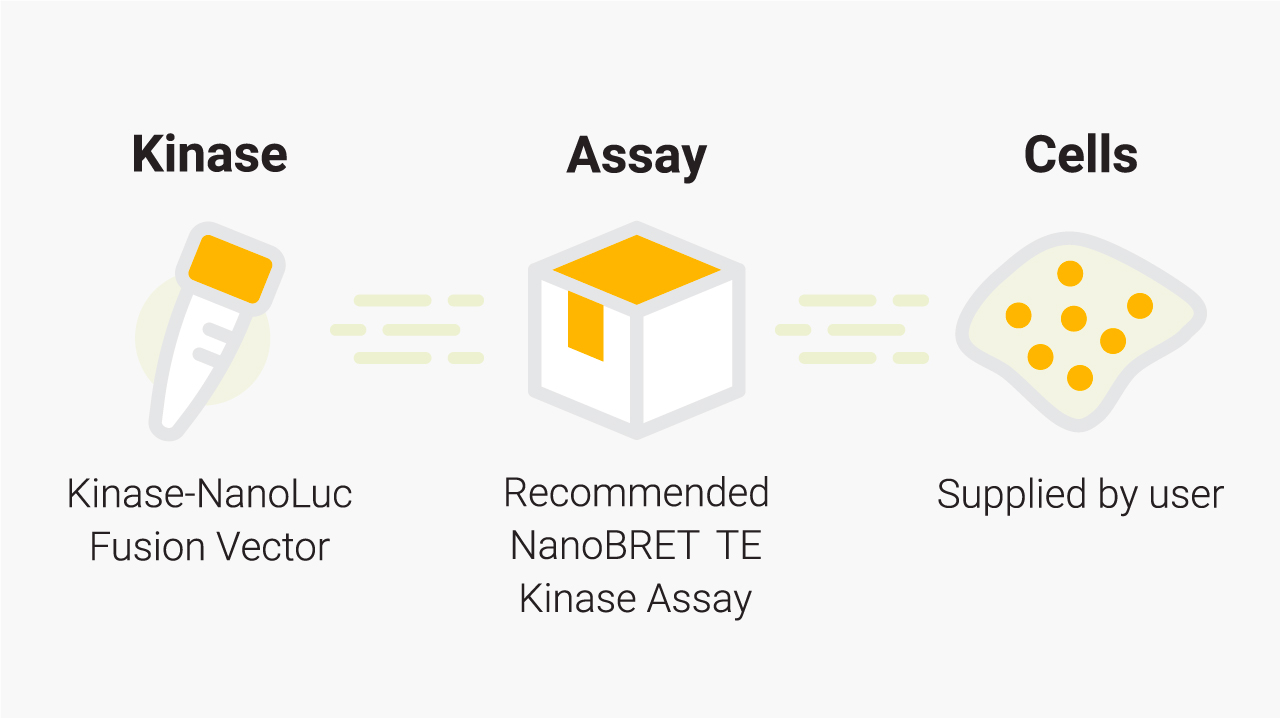
What you need for a NanoBRET® TE Kinase Assay. Promega supplies the individual Kinase-NanoLuc® fusion vector and the appropriate NanoBRET® TE Kinase Assay. For a given kinase, the recommended NanoBRET® TE Kinase Assay and application note can be found in the Kinase Target Engagement Selection Table. You will need to supply the cells and cell culture reagents. The tracers and substrate/inhibitor combinations are also available as standalone products.
A.

B.

Examining both cellular affinity and residence time in live cells can provide a more complete picture of a compound engaging a kinase. This example uses the NanoBRET® TE ABL1 kinase assay and several drugs targeting chronic myelogenous leukemia (CML). The first-generation CML drug Imatinib shows weaker binding (A) and shorter residence time (B) at ABL1, compared to second-generation (Dasatinib) and third-generation (Ponatinib) drugs. Ponatinib displays similar affinity (A) to Dasatinib but has much longer residence time (B).
Learn more about NanoBRET® Kinase Target Engagement Assays
Kinase Selectivity Profiling Systems
Biochemical Kinase Selectivity Profiling
The Kinase Selectivity Profiling Systems are optimized for fast and simple kinase profiling reactions. These systems include kinase and substrate pairs grouped in single kinase families, or as a general panel of kinases representative of the human kinome, for a broad kinase profile. They rely on the proven ADP-Glo™ Kinase Assay technology to make kinase profiling fast and simple. Systems can also be customized.
- Fast turnaround time in a matter of hours
- Flexible kinase inhibitory profiling
- Stable luminescent signal allows batch plate processing
Schematic Overview of the Kinase Selectivity Profiling System
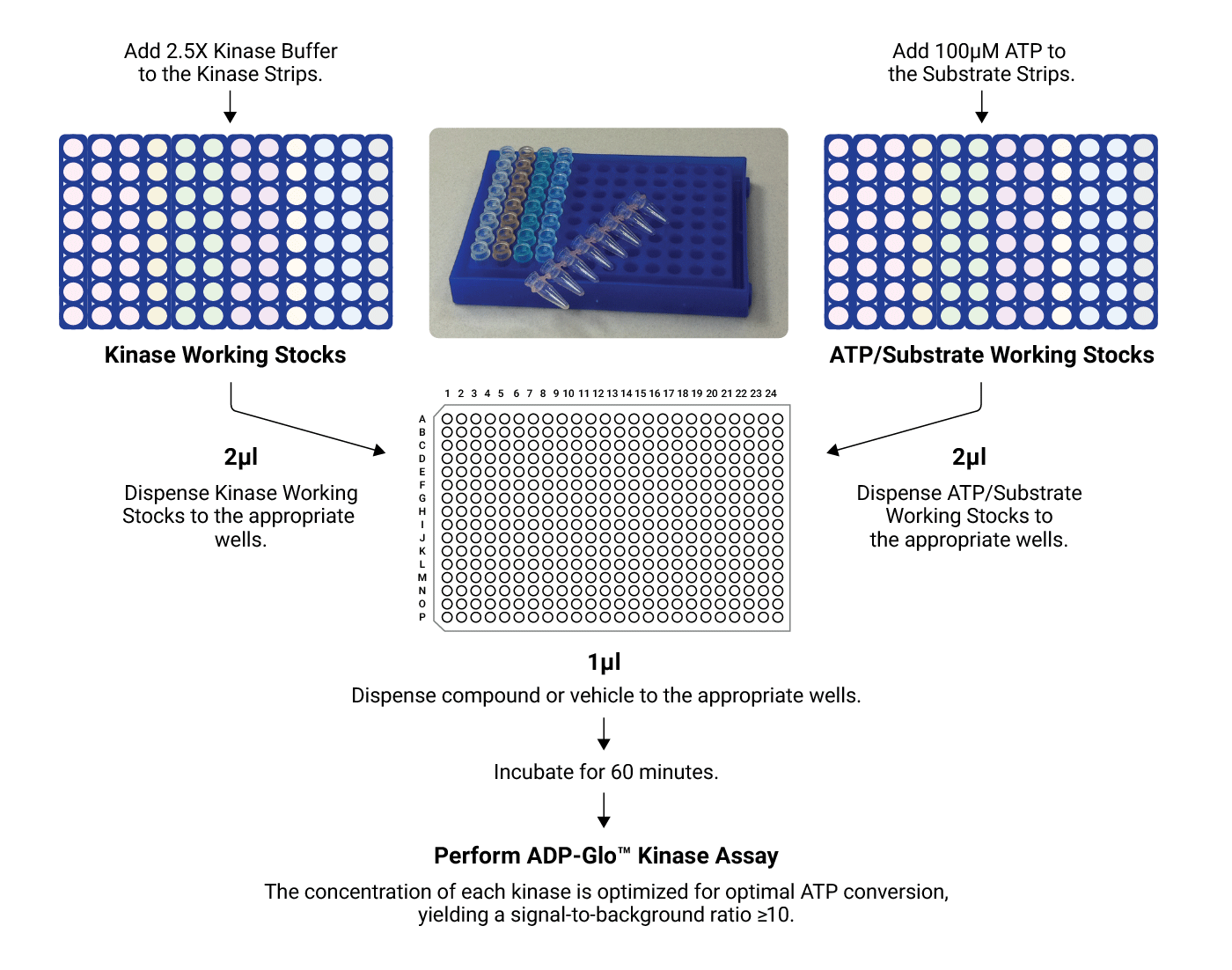
Concentrated kinases and substrates/cofactor stocks are first diluted and then combined with test compound in a 384-well plate. After a 1-hour incubation, the ADP-Glo™ Assay is performed. The resulting luminescent signal is proportional to ADP concentration and correlates with kinase activity.
Learn more about Biochemical Kinase Selectivity Profiling Products
Cellular Kinase Selectivity Profiling
Our new compound kinase cellular selectivity profiling service takes the advantages of NanoBRET® TE technology to quantitatively measure direct compound binding to kinases in live cells. Compounds can be tested at a single concentration against each kinase in a 192- or 240- kinase panel, transiently expressed in HEK293 cells. The output includes percent occupancy of test compounds against each kinase tested.
Kinase cellular selectivity profiling using NanoBRET® TE Intracellular Kinase Assays allows for:
- Uniquely enabling drug and chemical probe development.
- Better defining intracellular selectivity of a candidate compound and aiding drug repurposing via comprehensive profiling against a broad spectrum of kinases or kinase subfamilies.
Paper: Single tracer-based protocol for broad-spectrum kinase profiling in live cells with NanoBRET.
Learn more about our kinase selectivity profiling services.

A.

B.

Generation of crizotinib cellular kinase selectivity profile. A: Percent occupancy of crizotinib against a selective set of kinases; B: Inter-assay correlation analysis of crizotinib target occupancy for two independent experimental replicates.
Contact us to submit a question or service request for our kinase profiling services.
Phosphorylation and Pathway Analysis
The Lumit® Immunoassay Cellular System is a sensitive luminescent antibody detection method to examine phosphorylation and study inhibitors that target specific nodes of the major kinase signaling pathways.
- Less interference from chemical compounds; only a luminometer is required
- Homogeneous “add and read” format amenable to high-throughput screening
- Endogenous substrates eliminate the need for cell engineering and assays are compatible with any cell type, including primary cells
- Sensitive detection allows for assays to be performed on a low number of cells
- No immunoprecipitation step needed to detect phosphorylated protein
- “Do it yourself” assay can be adapted to any pathways or targets of interest
Lumit® Immunoassay Cellular Systems Complete Assays are now available.

Principle of the Lumit® Immunoassay Cellular System. The LgBiT and SmBiT subunits of NanoBiT® luciferase are conjugated to a pair of secondary antibodies from two different species (anti-rabbit, anti-mouse or anti-goat). Cells are lysed using a Lumit®-compatible lysis buffer (digitonin), and the target protein is detected by adding an antibody mix containing two primary antibodies (user supplied) against the protein along with the SmBiT- and LgBiT-conjugated secondary antibodies. Binding of the primary/Lumit® secondary antibody complexes to their corresponding epitopes brings the NanoBiT® subunits into close proximity to form an active NanoLuc® luciferase enzyme that produces light in proportion to the amount of the target protein.
View all Lumit® Immunoassay Cellular Systems
Kinase Biology Resources

Video: Automating the Kinase Profiling Systems
Learn how to use Promega kinase assays for rapid screening of compound libraries.
Poster: ADP-Glo: A Luminescent ADP Detection Assay for Kinases
In vitro studies of the mode of action of kinase inhibitors, and drug resistance mechanisms of mutated kinases.

Webinar: A Quantitative Technique to Measure Kinase Target Engagement in Live Cells
In this webinar we report the use of an energy transfer technique (NanoBRET) in the first quantitative approach to profiling kinase occupancy of a test compound inside live cells.