Target Degradation
Quantitative analysis of protein degrader function not only informs drug discovery and development but also enhances our understanding of cellular processes, enabling the development of more effective and targeted therapies while advancing our knowledge of biology and disease. HiBiT technology allows highly sensitive and real-time quantification of protein degradation, making it a powerful tool for high-throughput screening and analysis.
Read this whitepaper about studying protein degradation in real time using CRISPR knock-in cell lines and clones.

Is My Target Degraded?
HiBiT technology enables quantitative analysis of protein degrader function. HiBiT is an 11-amino-acid peptide tag, which has high affinity for its complementary partner, LgBiT. Together, they form a binary luminescent protein. Upon HiBiT-LgBiT engagement, the active luciferase protein produces a very bright and highly sensitive readout correlated to the endogenous target protein level, when HiBiT is introduced at the endogenous locus using CRISPR gene editing. The HiBiT peptide tag itself can be detected with Anti-HiBiT mAb, expanding the detection options for HiBiT-tagged proteins.
Addition of compounds that elicit degradation results in loss of luminescence signal, which is highly quantitative and can be measured in real time. Cellular dose-response curves can be obtained and monitored over a 24- to 48-hour time frame, allowing for accurate determination of degradation rate, Dmax, DC50 values and protein recovery.
This approach allows rapid rank ordering of degradation against a series of different parameters, and the assay is suitable for high-throughput screening.

An illustration of HiBiT fusion:LgBiT complementation inside a cell.



Degradation kinetics of endogenous HiBiT-BRD4 following PROTAC treatment. HiBiT was inserted at the endogenous BRD4 locus in the HEK293 LgBiT Cell Line. Cells were treated with a titration of MZ1 in CO2-independent medium containing Nano-Glo® Endurazine™ Substrate. Panel A. Kinetic luminescence; Panel B. Degradation rate; C: Dmax.
Study protein degradation in real time using CRISPR knock-in cell lines and clones.
Learn how in this white paper.
Is My Target Degraded in Native (Non-Modified) Cells?
We offer cellular methods to monitor degradation of a target protein in native cells. In this example, the Lumit® Immunoassay Cellular System is used to detect SMARCA 4 degradation.
Monitoring Targeted Protein Degradation with Lumit® Cellular Immunoassay

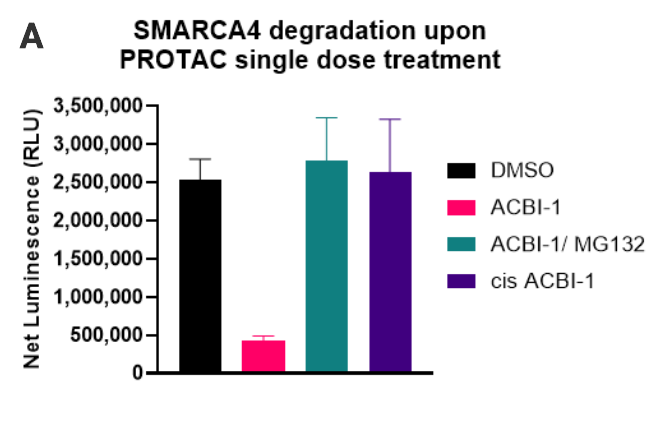
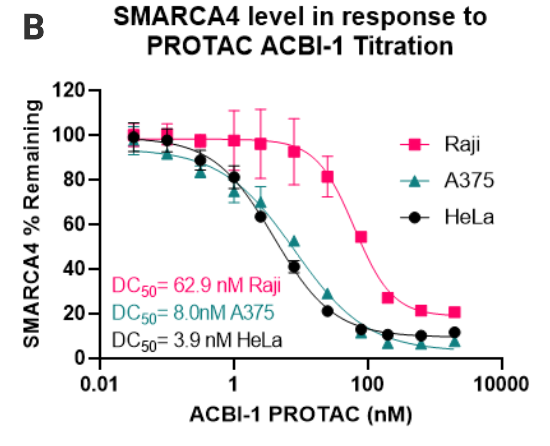
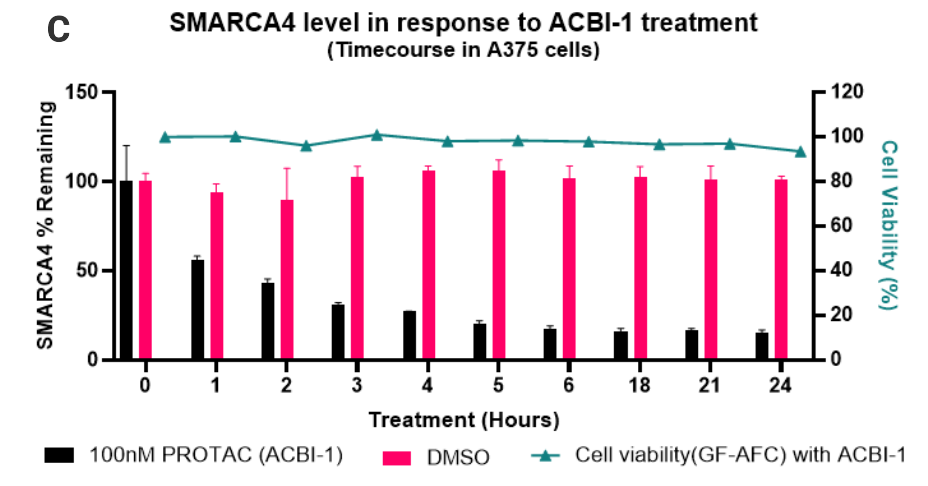
Degradation of native SMARCA4 following PROTAC treatment. 50,000 cells were seeded per well. After treatment with the indicated compounds, the Lumit® Immunoassay Cellular System (Set 2) was used to detect the level of SMARCA4. Panel A. SMARCA4 level after treatment with 250nM of ACBI-1, 250nM ACBI-1and 20µM MG132, 250nM Cis-ACBI-1, or DMSO for 5 hours. Panel B. Profiling SMARCA4 degradation with ACBI-1 in three different cell lines. Panel C. Time course of SMARCA4 degradation with ACBI-1.
Learn more about the Lumit® Immunoassay Cellular System.
Need help with Endpoint or Kinetic Degradation Profiling?
Learn more about our targeted protein degradation services.