SARS-CoV-2 Drug Discovery and Vaccine Development
Through innovation and collaboration, the scientific community rapidly came together to discover and develop numerous SARS-CoV-2 therapeutics and vaccines; however, the search for effective therapeutics and vaccines continues. The viral life cycle provides many areas that new drugs can target, from virus:host interaction to viral protease activity. Evolving SARS-CoV-2 variants require therapeutics and vaccines to adapt.
Promega offers several assays and enabling technologies for SARS-CoV-2 drug discovery and vaccine development. These assays and technologies include monitoring viral entry, detecting neutralizing antibodies and their ADCC or ADCP activity, and measuring viral protease activity.
Interested in how our technologies can help you with SARS-CoV-2 drug discovery and vaccine development? We can customize a virtual presentation to answer your questions.
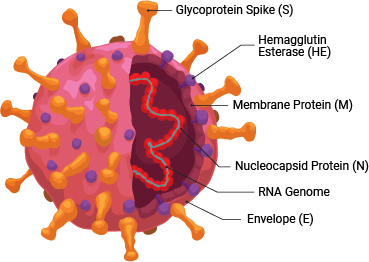
SARS-CoV-2 is a member of the coronavirus family. This retrovirus binds to the ACE2 host cell receptor through the viral surface Spike protein. The virus and its life cycle provide numerous opportunities for targeted drug discovery:
- Inhibiting the 3CLPro and PLPro viral proteases, which are necessary for the viral life cycle
- Preventing the binding of the viral Spike protein to the ACE2 host cell surface receptor, thereby preventing viral entry
- Modulating the effects of the hyperactivated immune system caused by SARS-CoV-2 infection, like inflammation and cytokine release
As with therapeutics, many vaccines in development target the host antibody response against the SARS-CoV-2 Spike protein, to prevent viral entry into host cells.
Learn more about products and technologies for viral research.
SARS-CoV-2 enters mammalian cells through binding of its Spike protein to the angiotensin-converting enzyme 2 (ACE2) receptor. Therapeutics that inhibit this interaction can prevent viral entry. The Lumit™ SARS-CoV-2 Spike RBD:hACE2 Immunossay measures the interaction between SARS-CoV-2 Spike protein receptor-binding domain (RBD) and human ACE2.

Lumit™ SARS-CoV 2 Spike RBD: hACE2 Immunoassay principle. Detection of RBD-rabbit Fc:hACE2-mouse Fc interaction is performed by Lumit™ secondary antibodies (Lumit™ Anti-Mouse Ab-LgBiT and Lumit™ Anti-Rabbit Ab-SmBiT). Panel A. Binding of the antibodies to the proteins promotes the reconstitution of NanoLuc® luciferase, generating bioluminescence. Panel B. The presence of a molecule that disrupts the RBD-hACE2 interaction reduces the bioluminescent signal.

Anti SARS-CoV-2 antibody screening using the Lumit™ SARS-CoV-2 Spike RBD:ACE2 Immunoassay detects neutralizing antibodies (Nabs) with different inhibitory profiles. Monoclonal antibodies were serially diluted and incubated with SARS-CoV-2 Spike RBD-rabbit Fc for 30 minutes prior to the addition of hACE2-mouse Fc and Lumit™ antibodies.

Learn more about the assay in this publication:
Alves, J. et al. (2021) A bioluminescent and homogeneous SARS-CoV-2 spike RBD and hACE2 interaction assay for antiviral screening and monitoring patient neutralizing antibody levels. Sci. Rep. 11, 18428.
Major SARS-CoV-2 Spike RBD mutant variants are also available for use in this assay to explore the influence of RBD mutations on antibody binding.
Contact us for more information on the Lumit™ SARS-CoV-2 Spike RBD:ACE2 Immunoassay, or visit the product page.
Using pseudotyped virus-like particles (PsVLPs), the SARS-CoV-2 HiBiT-PsVLP Assay was developed to measure the activity of small molecule inhibitors and neutralizing antibodies that block viral entry into the host cell. The assay uses innovative HiBiT bioluminescence technology to tag PsVLPs that contain the Spike protein on their surface. The PsVLPs are noninfectious because they contain no viral genetic material.
Assay Design
- HiBiT-tagged VLPs pseudotyped with SARS-CoV-2 Spike protein are added to SARS-CoV-2 Target Cells. HiBiT is packaged inside the PsVLPs
- In the absence of inhibitors or neutralizing antibodies (nAbs), SARS-CoV-2 HiBiT-PsVLPs bind to target cells via Spike/ACE2 interaction and undergo membrane fusion mediated by cellular proteases. HiBiT is released into target cells and binds to LgBiT to generate a luminescent signal in the presence of substrate.
- In the presence of inhibitors or nAbs against SARS-CoV-2 entry, the entry/fusion processes of PsVLPs are blocked, thereby preventing HiBiT release. No luminescent signal is produced.

Assay Workflow
This real-time assay is simple to use and can be completed in 3 hours. Advantages of this workflow include:
- Biologically relevant
- Quantitative
- Reduced biosafety requirements (BSL1)
- Simpler and faster than existing bioassays
- Add-mix-read format
- Turnaround time ≤8 hours - Convenience of Thaw-and-Use format
- No need to generate virus or culture cells

Assay Sensitivity





HiBiT-PsVLP Assay recapitulates differing sensitivity of SARS-CoV-2 variants to neutralization by therapeutic mAbs. SARS-CoV-2 LgBiT/Target Cells were incubated with various SARS-CoV-2 Spike neutralizing antibodies and each of the SARS-CoV-2 S HiBiT-PsVLPs carrying different spike variants (G614, Alpha, Beta, Gamma, and Delta). After 3 hours of incubation, Nano-Glo® Live Cell Reagent was added and luminescence was determined in a Glo-Max® Discover Plate Reader. Imdevimab (panel B) maintains neutralizing activity against all variants while the other three Nabs tested showed differential neutralizing activities against different variants (panels A, C and D).
Assay Reproducibility


HiBiT-PsVLP Assay shows excellent day-to-day reproducibility. SARS-CoV-2 LgBiT/Target Cells were incubated with SARS-CoV-2 Spike neutralizing antibodies and the SARS-CoV-2 S (G614) HiBiT-PsVLPs in three independent experiments on different days.
Contact us for more information on the SARS-CoV-2 HiBiT-PsVLP Assay, including assays available and in development for SARS-CoV-2 variants and other viruses.
The binding of SARS-CoV-2 Spike protein to the ACE2 receptor is a critical step for viral entry; thus, the Spike protein is a main target for antiviral drug discovery and vaccine development. Besides neutralization by blocking Spike:ACE2 interactions, anti-Spike antibodies may have additional antiviral activities mediated by the antibody Fc domain. These activities include antibody-dependent cellular cytotoxicity (ADCC) and antibody-dependent cellular phagocytosis (ADCP).
ADCC Reporter Assay
The FcγR Reporter Bioassay uses engineered reporter effector cells and engineered target cells stably expressing the SARS-CoV-2 Spike (S) protein:
- Anti-S antibody simultaneously binds to S protein on the target cells and the FcγRIIIa receptor on the ADCC Bioassay Effector Cells
- This binding leads to the activation of FcγRIIIa receptor and activation of luciferase expression in the ADCC Bioassay Effector Cells.



Measurement of ADCC reporter activity from Anti-SARS-CoV-2 S2 antibody. ADCC Bioassay Effector cells were incubated with SARS-CoV-2 S CHO-K1 target cells and anti-SARS-CoV-2 S2 antibody (Sino Biological, Cat.# 40590-D001) for 6 hours. Luminescence was measured using the Glo-Max® plate reader.
Contact us for more information on the SARS-CoV-2 ADCC Reporter Assay.
PBMC ADCC Reporter Assay
The PBMC ADCC Assay uses primary, ADCC-qualified PBMCs and engineered target cells stably expressing SARS-CoV-2 Spike (S) protein and a HaloTag®-HiBiT protein. The assay takes advantage of HiBiT extracellular complementation technology:
- Anti-S antibody simultaneously binds to S protein on target cells and FcγRIIIa on primary NK effector cells.
- When ADCC occurs, lysis of the target cells releases HiBiT protein into the medium, which can be detected by NanoGlo® HiBiT Extracellular Detection Reagent.




Measurement of ADCC activity from Anti-SARS-CoV-2 S2 antibody using primary cells. Primary PBMCs were incubated with SARS-CoV-2 S CHO-K1/HiBiT target cells and anti-SARS-CoV-2 S2 antibody (Sino Biological, Cat.# 40590-D001) for 5 hours. Luminescence was measured using the Glo-Max® plate reader.
Learn more about the SARS-CoV-2 PBMC ADCC Assay, featured in a Science publication, in this blog post.
The SARS-CoV-2 genome encodes two proteases of therapeutic interest. 3CLpro (Main Protease or Mpro) is a chymotrypsin-like protease, and PLpro is a papain-like protease. They cleave viral polyproteins into individual proteins, and thereby play an essential role in the lifecycle of the virus. These luminescent protease assays are designed to detect viral protease activity and screen for potential inhibitors.
Assay Design
Protease and test compound or vehicle are combined in an opaque white multi-well plate and a reaction is initiated by addition of a peptide-aminoluciferin (aLuc) substrate. The reaction is stopped, and luminescence is initiated by adding Luciferin Detection Reagent. Signal is recorded on a plate-reading luminometer. Inhibitors are identified as test compounds that reduce light output.
See this application note for a complete assay protocol.


Contact us for more information about 3CLpro and PLpro Luminescent Protease Assays.