Protein Interaction Analysis
Understanding which cellular molecules interact can help define protein pathways and recognition sites involved in gene expression. The HaloTag® Technology tool kit can capture both direct and indirect protein interactions as well as defining DNA binding sites and examining protein binding in live cells.
Characterize Binary and Higher Order Protein Complexes
Protein:protein interactions play critical roles in cellular processes, including replication, transcription, translation and signal transduction. The HaloTag® Mammalian Pull-Down Systems are designed to capture and purify intracellular binary and higher order protein complexes, including transient or weakly interacting partners. The covalent binding of the HaloTag® fusion bait protein means capturing the cellular protein complexes formed and identifying more physiologically relevant protein partners without interference from nonspecific binding.
Overview of the HaloTag® Mammalian Pull-Down System

Capture Interacting Proteins for Investigation
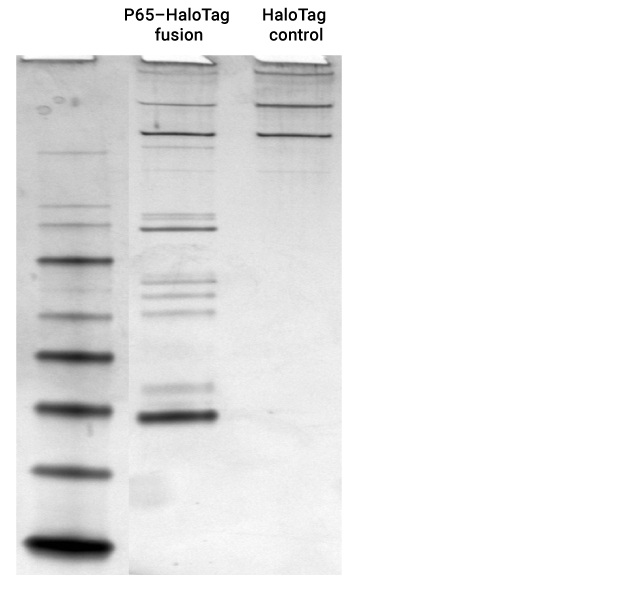
Expected cytoplasmic binary and tertiary protein interactions in the NFκB pathway were identified with specific p65-HaloTag® pull-down proteins identified by mass spectrometry were RelA(p65), RelB, C-Rel, IκBa, IκBb, IκBe, p100, p105(p50) and p52.
Study Protein:Protein Interactions in Live Cells
Investigating dynamics of protein interactions can be challenging. Both induced and inhibited protein:protein interactions can be studied in real time under cellular conditions using the NanoBRET® PPI Assay. Based on bioluminescence resonance energy transfer (BRET) technique where a bioluminescent donor protein brought into close proximity to a fluorescently tagged acceptor protein, the NanoBRET® Assay harnesses the bright NanoLuc® luciferase as the energy donor and the HaloTag® protein labeled with a fluorophore as the energy acceptor. This sensitive PPI assay means you can study full-length proteins expressed at low levels.

Depiction of energy transfer from a NanoLuc®-Protein A fusion (energy donor) to a fluorescently labeled HaloTag®-Protein B fusion (energy acceptor) upon interaction of Protein A and Protein B.
Additional HaloTag® Resources
Find Multiprotein Complexes in Cells
Learn about the localization of multiprotein Mediator complexes as well as isolating and characterizing their function in this poster.
Visualize Protein:Protein Interactions
Read this blog post to learn how the power of BRET can peer inside the cell to quantify, characterize and show proteins interacting.

Use a More Sensitive BRET Assay
See why the NanoBRET® Assay is a great tool for shedding light on protein:protein interactions in this blog post.
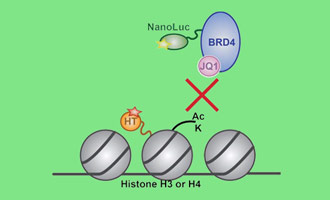
Measuring Drug-Target Interactions
This webinar describes how the NanoBRET® Assay can probe how possible modulators act on epigenetic proteins in live cells.