Quick and Easy Isolation of Genomic DNA from Drosophila Using the Maxwell® 16 Instrument
1Brown University, Providence, RI; 2Promega Corporation
Publication Date: 2010
Abstract
Maxwell® 16 Tissue DNA Purification Kit successfully purifies genomic DNA suitable for routine PCR genotyping and Southern blot analysis from whole flies, larvae, pupae, and other tissues. The simple and functional design allows Drosophila researchers to drop a fly into the cartridge, turn on the instrument, and walk away.
Introduction
DNA purification from Drosophila melanogaster typically requires time-consuming manual homogenization as well as careful preparation and handling of multiple reagents and buffers. Variability in overall DNA quality due to an individual's technique and expertise can compromise the results of important downstream applications. Automating this routine laboratory task improves consistency and frees up valuable researcher time and talent to accelerate the discovery process. Here we report the utility of Maxwell® 16 to automate Drosophila DNA purification from whole animals and body parts for genotyping by PCR and Southern blot.
"The mechanical action of the Maxwell® 16 plunger breaks apart many sample types, allowing fully automated extraction without the need for preprocessing."
The Maxwell® 16 System combines compact instrumentation, prefilled reagent cartridges and optimized automated methods to perform DNA purification of 1–16 samples with minimum handling. The system uses MagneSil® paramagnetic particles to bind nucleic acids and moves the particles through a series of wash steps prior to elution of the purified sample. This technology has been adapted to a range of purification technologies for DNA, RNA and protein, making Maxwell® 16 a highly versatile, integral tool in the laboratory. The mechanical action of the Maxwell® 16 plunger breaks apart many sample types, allowing fully automated extraction without the need for preprocessing—a particular advantage to Drosophila researchers.
Early and Non-Lethal Drosophila Genotyping
When conducting a large genetic screen, early detection of animals and populations that do not carry the desired genotype can save a researcher considerable time and money in terms of manual labor and maintenance materials (food, plastics, etc.) As a proof-of-concept experiment, DNA was isolated from various sample types from the same population of animals using the Maxwell® 16 Tissue DNA Purification Kit (Cat.# AS1030; Figure 1). Samples were placed directly into the buffer in the first well of each cartridge, and 300µl of water was added to the elution tubes. Cartridges were loaded into the instrument and run on the DNA Tissue method (~45 minutes) according to the standard protocol.
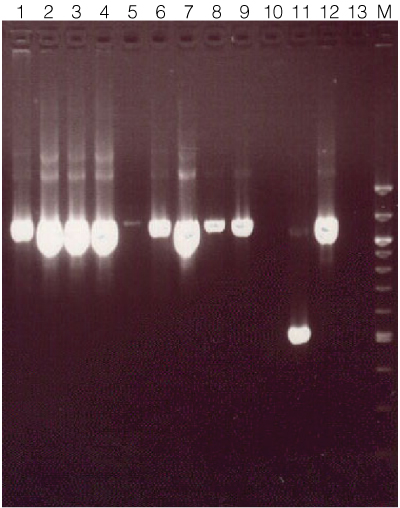 Figure 1. Amplification results using genomic DNA isolated from various Drosophila sample types.
Figure 1. Amplification results using genomic DNA isolated from various Drosophila sample types.
Genomic DNA was isolated using the Maxwell® 16 Tissue DNA Purification System from the following Drosphila tissue types: Lane 1, 1 whole fly; lane 2, 12 fly heads; lane 3, 1 fly head; lane 4, 1 whole fly; lane 5, 8 pairs of fly testis; lane 6, ~60mg 3rd instar larvae; lane 7, ~60mg pupae; lane 8, 12 pairs of fly wings; lane 9, 12 pairs of fly wings (frozen at –80°C for 5 minutes prior to processing)*; lane 10, blank; lane 11, positive control (~550bp); lane 12, positive control (~1,100bp); lane 13, blank; lane M, 100bp ladder (New England Biolabs). Amplification was performed using either 1µl (lanes 1–4, 8–9) or 5µl of a 1:100 dilution of eluate (lanes 5–7) as the template. DNA concentration was not measured for this experiment. Amplification products were separated on a 1.5% agarose gel. *When DNA was measured by spectrophotometer (data not shown), freezing whole animals, pupae, and wings at –80°C for 5 minutes prior to processing typically resulted in higher yields.
Detection of Transgenic Homologous Recombination (HR) Construct
Homologous recombination is a powerful technique that allows Drosophila researchers to introduce specific sequence mutations directly into a gene of interest. The targeting construct is introduced into Drosophila by standard microinjection techniques. The construct consists of two ~2.5 kb homology arms containing the desired mutation(s) and a selectable eye color marker (1) (2) . These constructs also contain Flp recombinase and I-SceI endonuclease sites that allow for excision of the transgene and Cre/loxP sites that allow for removal of the eye color marker after integration. The excised transgene acts as an extrachromosomal donor that recombines with the gene of interest via the homology arms and is followed via the eye color marker. Once integration is confirmed, the eye color marker is removed via Cre recombinase acting on lox P sites that flank the eye color marker.
While the homologous recombination technique has a high success rate, there can be a high background of false positives at the initial integration step. To rapidly identify successful integrations, a validation PCR is performed on individual candidate flies after mating success is confirmed. Separate PCR amplifications are run to validate both arm 1 and arm 2 (Figure 2). Lines that validate (Figure 2, Panel A) are expanded and undergo a series of crosses to remove the eye color marker via Cre recombinase. After removal of the eye color marker, lines carrying the integrated construct are determined by PCR. The primer pair used for this step will amplify the same size product from both endogenous and recombinant loci (Figure 2, Panel B). The two products can be distinguished by presence or absence of the Acc65I restriction site that is present only in the recombinant locus (Figure 2, Panel C). Lines that fail to validate with both arms can be discarded.
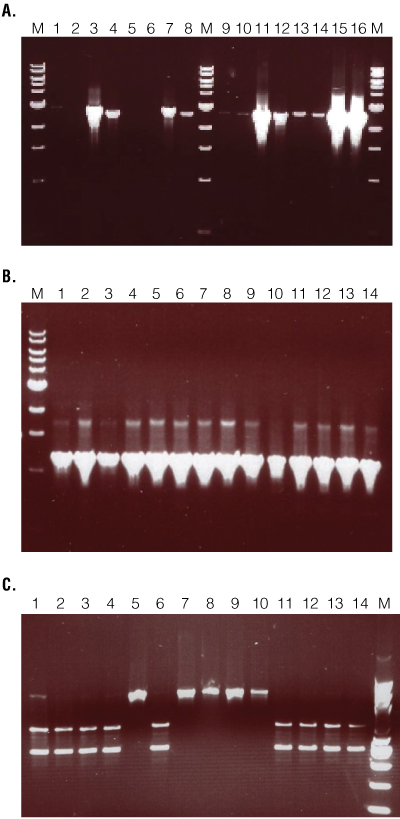 Figure 2. Validation PCR results for recombinant construct integration.
Figure 2. Validation PCR results for recombinant construct integration.
DNA was isolated from individual flies using the Maxwell® 16 Tissue DNA Purification Kit and eluted in 300µl of water. One to five microliters of eluate was amplified 40 cycles in a 50µl reaction using 0.25µl GoTaq® DNA Polymerase (Promega). The primers used were complementary to the vector sequence of a construct designed for a homologous recombination (HR) experiment and the endogenous sequence. Ten microliters of PCR product was separated on a 0.8% (Panels A and B) or 1.5% (Panel C) agarose gel. Panel A. Validation of targeting events. Two primer pairs for each arm were used to validate three samples: A, B, and C. Lanes 2 and 3, Arm 1 sample A; lanes 4 and 5, Arm 1 sample B; lanes 6 and 7, Arm 1 sample C; lanes 8 and 9, Arm 1 positive control; lane 11 and 12, Arm 2 sample A; lanes 13 and 14, Arm 2 sample B; lanes 15 and 16, Arm 2 sample C; lanes 17 and 18, Arm 2 positive control. Lanes M, 1kb ladder (NEB). Only sample B produced the expected amplification product with both arms and both primer pairs. Panel B. Post-Cre amplification of junction region. Successful integration of homology arms and subsequent Cre removal of the mini-white gene (eye color) results in 75 bases of vector sequence replacing endogenous sequence between arms 1 and 2. Primers designed to arms 1 and 2, flanking the 75 bases of intervening sequence, amplify product from both endogenous and recombinant loci. The expected 1,197bp sequence was successfully amplified in all samples. Panel C. Restriction digest to distinguish endogenous from recombinant loci. The integrated sequence retains an Acc651 restriction site that can be used in a diagnostic digest to differentiate recombinant and endogenous loci. The samples in lanes 1–4, 6 and 11–14 yielded the expected restriction digest fragment.
Screening Wild Populations for a Repetitive Element by Southern Blot Analysis
Southern blot analysis remains a standard method for detecting multiple-copy DNA sequences such as integrated transgenes, microsatellite markers and repetitive elements (3) . Here, a collection of wild-caught Drosophila populations from diverse geographic locations was analyzed for presence and quantity of a repetitive element (Rsp) (Figure 3).
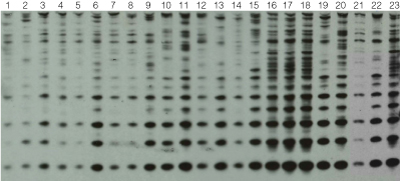 Figure 3. Southern Blot analysis for the presence of the Rsp repetitive element.
Figure 3. Southern Blot analysis for the presence of the Rsp repetitive element.
Twelve whole flies from each wild-caught population were processed using the Maxwell® 16 Tissue DNA Purification Kit. Three hundred nanograms of each DNA sample was digested with XbaI and separated on a 1.5% agarose gel. Known populations were used as "standards" (lanes 21, 22 and 23). The gel was blotted and probed with the Rsp element. Probe DNA was directly labeled with horseradish peroxidase (HRP) (GE Healthcare) and detected using chemiluminescence (Pierce).
Conclusion
The Maxwell® 16 Tissue DNA Purification Kit successfully purifies genomic DNA from whole flies, larvae, pupae, and other tissues that is suitable for routine PCR genotyping and Southern blot analysis. The simple and functional design allows Drosophila researchers to drop a fly a cartridge, turn on the instrument, and walk away. This frees up valuable time to accomplish other tasks.
Related Products
Related Resources
Article References
- Rong, Y.S. and Golic, K.G. (2000) Gene targeting by homologous recombination in Drosophila. Science 288, 2013–8.
- Engels, W.R. (2000) Genetics: Reversal of fortune for Drosophila geneticists? Science 288, 1973–5.
- Southern, E.M. (1975) Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98, 503–17.
How to Cite This Article
Scientific Style and Format, 7th edition, 2006
Staber, C. and Mann, R. Quick and Easy Isolation of Genomic DNA from Drosophila Using the Maxwell® 16 Instrument. [Internet] 2010. [cited: year, month, date]. Available from: https://www.promega.com/resources/pubhub/quick-and-easy-isolation-of-genomic-dna-from-drosophila-using-the-maxwell-16-instrument/
American Medical Association, Manual of Style, 10th edition, 2007
Staber, C. and Mann, R. Quick and Easy Isolation of Genomic DNA from Drosophila Using the Maxwell® 16 Instrument. Promega Corporation Web site. https://www.promega.com/resources/pubhub/quick-and-easy-isolation-of-genomic-dna-from-drosophila-using-the-maxwell-16-instrument/ Updated 2010. Accessed Month Day, Year.
GoTaq, MagneSil and Maxwell are trademarks of Promega Corporation.
Products may be covered by pending or issued patents or may have certain limitations. Please visit our web site for more information.