Designing a Bioluminescent Reporter Assay—Choosing Your Experimental Reporter
What Makes a Good Reporter Assay?
When it comes to designing a good reporter assay there are a number of parameters to consider: What cells should I work in? How will I introduce my reporter into my cells? How should I design my reporter construct? What controls should I use? Two of the fundamental elements you need to consider are the reporter protein itself and the assay chemistry used to detect reporter activity. Attributes of these two components together contribute strongly to the quality of the assay.
We use three luciferases as reporters in our bioluminescent assays: NanoLuc® Luciferase (19kDa), Renilla Luciferase (36kDa) and Firefly Luciferase (61kDa). Including the destabilized versions of Firefly and NanoLuc® Luciferases, we offer 5 different bioluminescent reporters and multiple detection assays for each. So, how do you choose the combination that is right for you? First, let's look at the reporter characteristics.
Ideal Characteristics of Reporter Assay Components
Reporter Protein:
- enzymatic activity (signal amplification)
- small
- no processing to be active
- monomer
Reporter Assay Chemistry:
- low background
- easy to use and measure
- quantitative
- sensitive, broad dynamic range
Signal Strength
NanoLuc® Luciferase provides the brightest signal. On a per mole basis, NanoLuc® Luciferase is approximately 100-fold brighter than Firefly or Renilla Luciferase. This difference in brightness can be seen in cells as well, and makes NanoLuc®-based assays more sensitive than those using other luciferases.
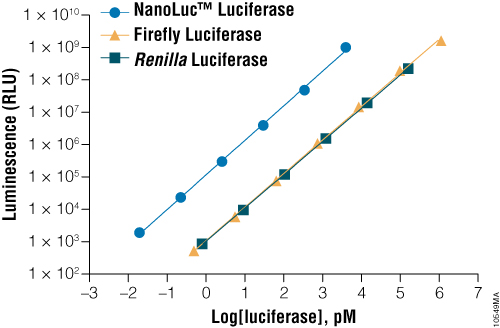
Figure 1. A comparison of the sensitivity of NanoLuc®, firefly and Renilla luciferase assays. Luminescence was measured from varying concentrations of purified proteins after mixing the reporter enzyme with its respective detection reagent. NanoLuc® luciferase was approximately 150-fold brighter than firefly or Renilla luciferases at equivalent concentrations. Detection reagents used were Nano-Glo® Luciferase Assay Reagent for NanoLuc® luciferase, ONE-Glo™ Luciferase Assay System Reagent for firefly luciferase and Renilla-Glo™ Luciferase Assay System Reagent for Renilla luciferase.
Response Time
NanoLuc (Nluc) and Firefly (luc2) Luciferases have destabilized versions (NlucP and luc2P) that contain the PEST protein degradation sequence, targeting them for rapid degradation. The PEST sequence drastically reduces the half-life of the enzyme. A destabilized reporter will have a smaller intracellular pool of protein allowing for a faster, more sensitive response to changes in expression.
Half-life varies between Nluc and Fluc reporters as follows:
- NlucP - 20 minutes
- Fluc2P - 1 hour
- Fluc2 - 3 hours
- Nluc - 6 hours
In the experiment shown below, constructs using an NFkB promoter and either a NlucP, FlucP, Nluc or Fluc reporter gene were used to transfect HEK293 cells. Cells were treated at time 0 with TNFα, which induces expression from the NFkB promoter. The fastest and strongest response was from the NlucP construct followed by the FlucP construct.
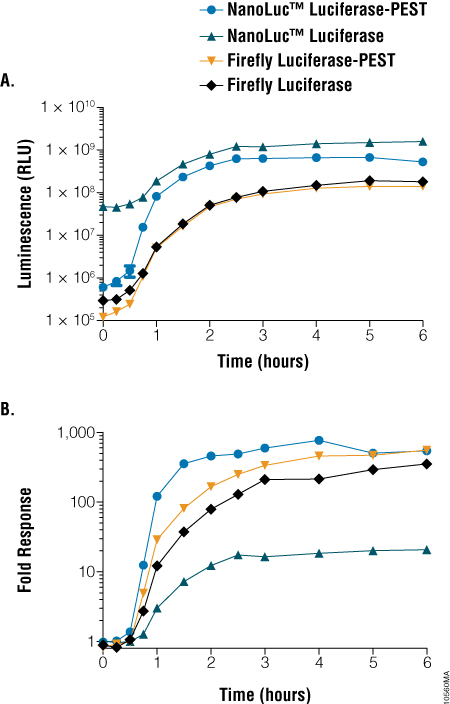
Figure 2. Comparison of light output and response dynamics between NanoLuc® and firefly luciferase constructs. HEK293 cells were transiently transfected with constructs encoding various forms of luciferase under the control of a minimal promoter containing five repeats of a consensus NF-κB response element and replicate plates treated with 100ng/ml of rhTNF-α for the indicated times. Panel A. Luminescence was measured at the indicated time points. Panel B. Fold induction was calculated by comparing treated samples to untreated samples at the same time point.
Sensitivity for Weak Responses
The previous experiment measured a strong response to a stimulus. Many physiological responses are more subtle. How would these four reporters perform in an assay measuring a weaker response? The experiment below shows that NlucP is the best choice when sensitivity is an issue. In these experiments, the heat-shock response element (HSE) was used to drive expression of each reporter. Treatment of the cells with the heat-shock mimetic, 17-AAG [17-(allylamino)-17-demethoxygeldanamycin), activates the hsf1 transcription factor which in turn activates the HSE. The NlucP reporter construct is the only one that demonstrates a significant response above background.

Choosing an Assay
Choosing a detection assay depends on several factors. As shown earlier, your choice of reporter will affect signal strength, response time and sensitivity. If you choose Nluc or NlucP as your reporter, you will detect activity with the Nano-Glo® Luciferase Assay System, which provides stable, glow-type signal in the presence of Nluc, with a half-life of approximately 120 minutes.
In many cases, firefly luciferase is still a very good reporter option and many constructs exist using Fluc and FlucP as the reporter. There are multiple assay chemistry options for detection of the Fluc reporter allowing you to choose the detection assay most appropriate for your particular experimental needs. Assays for detection of firefly luciferase include options with either “flash” or “glow” signal kinetics. A “flash” assay is one where the signal is strong but short-lived requiring injection-based delivery of reagent. In contrast, a “Glow” type assay has a prolonged signal eliminating the need for injectors, but with generally lower signal intensities. Learn more about “flash vs glow”. The figure below compares signal strength and duration using different firefly assays with glow signal kinetics. The Bright-Glo™ Assay provides the brightest signal, but it decays rapidly. On the other hand, the Steady-Glo® Assay provides the most consistent signal duration with the lowest intensity. The ONE-Glo™ and ONE-Glo™ EX assays are ideally suited for high-throughput use. The ONE-Glo™ EX Reagent has the added advantage of improved stability, allowing you to make up the reagent and store it at 4⁰C for several weeks, simplifying repeat use if you have a set of experiments to run.
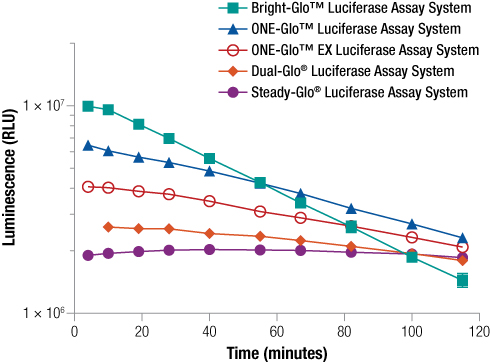
Figure 4. Comparison of five single glow kinetic firefly luciferase reporter assays. One hundred microliters of purified firefly luciferase (13.8ng/ml in DMEM with 0.1% Prionex® as carrier) was combined in a 96-well plate with 100µl of Bright-Glo™, ONE-Glo™, ONE-Glo™ EX, Dual-Glo® Luciferase or Steady-Glo® Reagents. Luminescence was measured periodically over 2 hours, n = 8.
What About Dual-Reporter® Assays
Dual-reporter® assays are an excellent way to control for variations in your results or maximize your data. Firefly and Renilla luciferases can be used together with the Dual-Luciferase® (flash kinetics) or Dual-Glo® (stable kinetics) Assay Systems. The choice largely depends on the sensitivity and signal stability required for your experiment. Alternatively, firefly and NanoLuc® luciferases can be combined in the Nano-Glo® Dual-Luciferase Reporter Assay (NanoDLR™) for improved sensitivity in dual-reporter assay with stable signal kinetics.
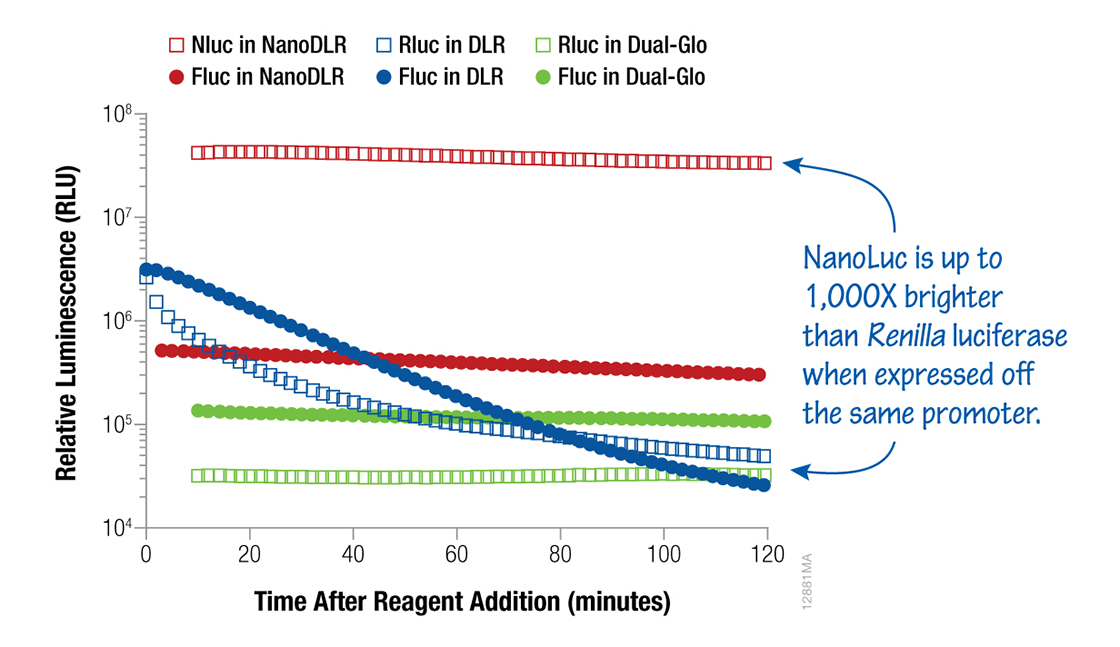
Figure 5. Dual-Luciferase® assays. Luminescence signals from HEK293 cells transfected with a 1:1:8 ratio of either TK-Rluc (Renilla):TK-Fluc (firefly):carrier DNA or TK-Nluc (NanoLuc® Luciferase):TK-Fluc:carrier DNA were assayed with NanoDLR™, DLR™ or Dual-Glo® Dual-Luciferase Assay System as indicated. When luciferases are expressed from the same promoter, the NanoDLR™ Assay provides bright, stable, “glow-type” signals for both Fluc and Nluc reporters.
Related Products and Resources
Further Learning
For a side-by-side comparison of Promega Firefly, NanoLuc® and Renilla reporter assays, see the Reporter Assay Comparison Guide. For detailed data and information on individual assays, visit the product page for each system.