The Latest and Greatest NIST PCR-Based DNA Profiling Standard: Updates and Status of Standard Reference Material® (SRM) 2391c
National Institute of Standards and Technology, Applied Genetics Group, Gaithersburg, Maryland, USA
Publication Date: 2011
Why use reference materials?
Since the late 1980s, the National Institute of Standards and Technology (NIST) has had scientists involved in DNA testing and over the past two decades has worked closely with the forensic community to develop DNA reference materials(1). In the years since the introduction of reference materials to the DNA human identity forensic community, the FBI has adopted these materials in their quality assurance standards. Specifically, under Section 9 on analytical procedures, FBI DNA Quality Assurance Standard 9.5.5 states, “The laboratory shall check its DNA procedures annually or whenever substantial changes are made to the protocol(s) against an appropriate and available NIST standard reference material or standard traceable to a NIST standard”(2).
Standard reference materials (SRMs) are prepared and qualified through careful characterization of component homogeneity, stability and reproducibility. A Report of Analysis (ROA) is created during preparation and characterization of the SRM materials. A master file is maintained by NIST scientists containing all of the values that go into developing the SRM components. The ROA is submitted and approved at the division level and by the Measurement Services Division (the office that is responsible for SRM sales). A Certificate of Analysis (COA) is prepared for each SRM describing the components and the certified values characterized for the SRM and is available to the general public.
Homogeneity, stability and reproducibility must be demonstrated on the materials used for the SRM. For example, SRM 2391 contains cell line 9948, which exhibits a tri-allelic pattern at CSF1PO. Different lots of 9948 used in SRM 2391a (1999) and SRM 2391b (2003) possess different levels of this tri-allelic pattern. It has been demonstrated that cell lines have exhibited different STR allele patterns after various passage numbers(3).
Note that the COA contains information regarding how long the SRM components should be stable. The actual materials contained within a NIST SRM may be purchased from an outside vendor, but characterization of the materials is typically conducted by NIST scientists. Interlaboratory studies are usually performed to verify homogeneity, stability and lab-to-lab reproducibility of the SRM component materials prior to their release for public purchase.
When new lots of SRM materials are created, comparisons are made at NIST to previous lots, but the final COA corresponds to the actual data obtained in characterizing the components present in the SRM lot being released.
What is in NIST SRM 2391c?
Standard Reference Material® 2391c (SRM 2391c) PCR-based DNA Profiling Standard is the third renewal of this SRM, originally released in 1995(4). SRM 2391c consists of six components labeled A through F. Components A through D are supplied as genomic DNA solutions packaged in perfluoroalkoxy fluoropolymer (PFA) vials (with component A as a single-source female; component B as a single-source male; component C as a single-source male; and component D as a mixture of 3 parts components A and 1 part component C). Components E (single-source female) and F (single-source male) are cells (≈ 75,000 cells per spot) deposited on 903 and FTA® papers, respectively, two 6mm punches per component. Inclusion of two kinds of storage paper will enable laboratories to test direct PCR methods.
In selection of DNA materials to be used, more than 25 different cell lines were screened to avoid tri-allelic patterns and <70% peak height ratio in heterozygous loci to select as many nonoverlapping alleles as possible and to provide similar quantitation values between currently available qPCR kits (Table 1). The DNA concentrations are given for information purposes only; these materials are not to be used as qPCR standards.
![[DNA] in ng/µl for Components A, B, C and D.](https://scv10mr-cdnpre-p-cus-00.azureedge.net/-/media/images/resources/tables/profiles-in-dna/1402/klinetable1.jpg?rev=c939ddb57e194171a4da846aaaec4da2&sc_lang=en) Table 1. [DNA] in ng/µl for Components A, B, C and D.
Table 1. [DNA] in ng/µl for Components A, B, C and D. The components of SRM 2391c are from different sources than those used in SRM 2391, SRM 2391a and SRM 2391b. Certified or Reference short tandem repeat (STR) genotypes for 68 loci (51 autosomal and 17 Y-STRs) across the six components are supplied in the Certificate of Analysis, which can be downloaded from the NIST web site. In order to avoid any potential null alleles, the SRM 2391c components were tested with 22 different commercial genotyping kits, which are listed in Table 2. There were no discordant results observed in the tested loci with these materials. In SRM 2391b, there was allele dropout in component 8 with allele 11 at D16S539 using the MiniFiler™ kit(5).
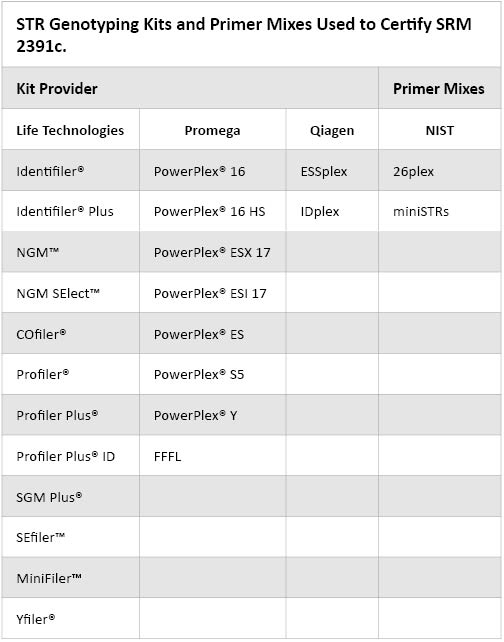 Table 2. STR Genotyping Kits and Primer Mixes Used to Certify SRM 2391c.
Table 2. STR Genotyping Kits and Primer Mixes Used to Certify SRM 2391c. What is the difference between Certified Values, Reference Values and Information Values?
A NIST Certified Value is a value for which NIST has the highest confidence in its accuracy in that all known or suspected sources of bias have been investigated or taken into account. In practical terms the loci of interest must be compared with multiple primer sets, or all alleles present for that locus must be sequenced to validate the genotype. The SRM is certified for genetic loci of forensic interest that were available in commercial STR typing kits at the time of production. The list for these certified loci can be found in Table 3.
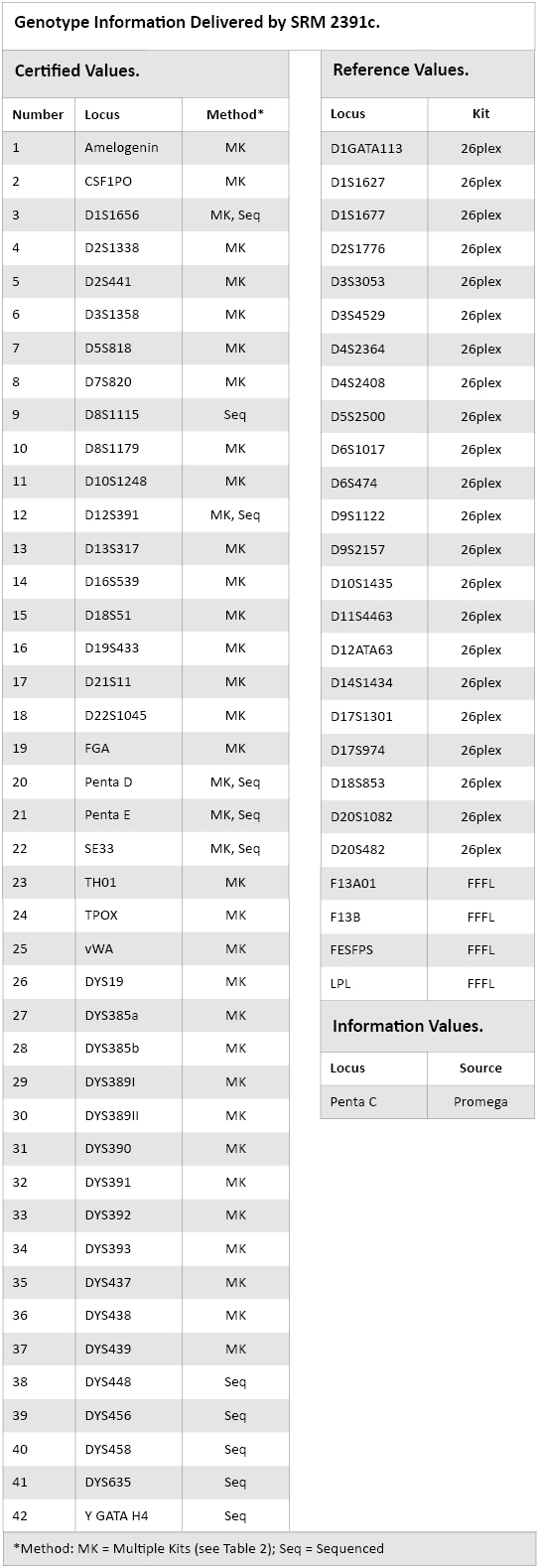 Table 3. Genotype Information Delivered by SRM 2391c.
Table 3. Genotype Information Delivered by SRM 2391c. A NIST Reference Value is a best estimate of the true value where all known or suspected sources of bias have not been fully investigated by NIST. Table 3 lists the loci that are considered reference values. Reference values are assigned when there is sequence information for only a few alleles of a particular locus. Alleles that do not have sequence information for the components of this SRM are considered reference values in terms of the repeat count based on base pair (bp) size differences of a nonsequenced allele compared to a sequenced allele for that locus.
A NIST Information Value is considered to be a value that may be of interest and use to the SRM user, but insufficient information is available to assess the uncertainty associated with the value. Information values have been assigned to the DNA concentrations of components A through D (Table 1) and genotypes for the Penta C locus (Table 3). For additional information regarding certified, reference and information values please see May et al.(6).
What is involved in the NIST certification process?
All components were tested with the kits listed in Table 2 and found to have concordant results for the loci shared across different genotyping kits. Electropherograms were closely examined for any indication of the presence of contamination, and none was found. Sequencing results confirmed the genotyping results obtained where additional weight to the genotyping data was either desired or needed for the level of certification as well as verification of genotypes of loci with only one primer set. Component D was analyzed at 1ng and 2ng of input DNA, and the peak heights and peak areas of unshared alleles in components A and C were used to evaluate the mixture ratio. Electropherograms for component D also were analyzed with TrueAllele® software, a mixture-deconvolution program from Cybergenetics (Pittsburgh, PA). All methods indicated the approximate 3-to-1 mixture had been accomplished.
The homogeneity of cells spotted on paper for components E and F was evaluated by selecting 70 spots from each component. Component E spots, composed of 903 paper, were extracted using the EZ1™ Advanced XL DNA Purification Robot and eluting in 100µl of TE–4 buffer. After quantification 1ng of the extracts were amplified with the PowerPlex® 16 HS System. All extracted spots produced acceptable electropherograms.
Component F spots, composed of FTA® paper, were subsampled with a 1.2mm punch. These subsamples were washed following the Whatman FTA® protocol BD01. The washed FTA® spots were amplified with PowerPlex® 16 HS. All but one of the 1.2mm spots produced acceptable electropherograms. The one spot that failed was resampled and was amplified successfully with Identifiler® Direct.
Four laboratories assisted in the interlaboratory assessment of the SRM (Palm Beach Sheriff’s Office, Promega Corporation, Life Technologies and Bode Technology Group). All genotypes generated were concordant.
What are the major differences compared to previous SRM 2391 series components?
All materials used are different from previous NIST SRMs. Thus, no 9947A and 9948 samples are present in SRM 2391c. There are six components in the current SRM 2391c compared to 12 in SRM 2391b. Different tube types (PFA vs. polypropylene) are used to aid improved recovery of materials. A two-person mixture is included with its individual components being present in the other genomic DNA samples. Different cells have been supplied on 903 and FTA® papers to enable DNA extraction or direct PCR methodologies. Finally, all male components are certified for 17 Y-STRs.
Acknowledgments and Disclaimers
We thank David Duewer, NIST Analytical Chemistry Division, for his invaluable input into NIST SRM development and statistical analysis. This work was funded in part by the National Institute of Justice (NIJ) through an interagency agreement 2008-DN-R-121 with the NIST Office of Law Enforcement Standards. Points of view in this document are those of the authors and do not necessarily represent the official position or policies of the U.S. Department of Justice. Certain commercial equipment, instruments and materials are identified in order to specify experimental procedures as completely as possible. In no case does such identification imply a recommendation or endorsement by the National Institute of Standards and Technology nor does it imply that any of the materials, instruments or equipment identified are necessarily the best available for the purpose.
Article References
- Reeder, D.J. (1999) Impact of DNA typing on standards and practice in the forensic community. Arch. Pathol. Lab. Med. 123, 1063–5.
- (2011) FBI Quality Assurance Standards for Forensic DNA Testing Laboratories.
- Parson, W. et al. (2005) Cancer cell line identification by short tandem repeat profiling: Power and limitations. FASEB J. 19, 434–6.
- About NIST Standard Reference Materials®, SRM. NIST web site.
- Kline, M.C. and J.M. Butler (2008) Understanding “null” alleles and STR allele mobility issues through variant allele sequencing. Presented at the American Academy of Forensic Sciences 2008 Meeting, February 23, 2008.
- May, W.E. et al. (2000) Definitions of terms and modes used at NIST for value-assignment of reference materials for chemical measurements. NIST Special Publication 260-136.
How to Cite This Article
Scientific Style and Format, 7th edition, 2006
Kline, M.C. et al. The Latest and Greatest NIST PCR-Based DNA Profiling Standard: Updates and Status of Standard Reference Material® (SRM) 2391c. [Internet] 2011. [cited: year, month, date]. Available from: https://www.promega.com/resources/profiles-in-dna/2011/the-latest-and-greatest-nist-pcr-based-dna-profiling-standard/
American Medical Association, Manual of Style, 10th edition, 2007
Kline, M.C. et al. The Latest and Greatest NIST PCR-Based DNA Profiling Standard: Updates and Status of Standard Reference Material® (SRM) 2391c. Promega Corporation Web site. https://www.promega.com/resources/profiles-in-dna/2011/the-latest-and-greatest-nist-pcr-based-dna-profiling-standard/ Updated 2011. Accessed Month Day, Year.
Contribution of an article to Profiles in DNA does not constitute an endorsement of Promega products.
Plexor and PowerPlex are registered trademarks of Promega Corporation.
COfiler, Identifiler, Profiler, Quantifiler, SGM Plus and Yfiler are registered trademarks of Life Technologies. EZ1 is a trademark of Qiagen GmbH Corporation. FTA is a registered trademark of Flinders Technologies, Pty, Ltd., and is licensed to Whatman. MiniFiler, NGM, NGM SElect, Profiler Plus and SEfiler are trademarks of Life Technologies. Standard Reference Material is a registered trademark of the National Institute of Standards and Technology. TrueAllele is a registered trademark of Cybergenetics, Inc.