Using Endoproteinases Asp-N and Glu-C to Improve Protein Characterization
Laurie Engel1, Sergei Saveliev1, Marjeta Urh1, Dan Simpson1, Richard Jones2 and Keith Wood1
1Promega Corporation, 2MS Bioworks
Abstract
Two alternative proteinases, Asp-N Sequencing Grade and Glu-C Sequencing Grade are available from Promega to expand options for protein digestion. Asp-N, Sequencing Grade, is a metallo proteinases that preferentially cleaves at the N-terminus of aspartic and cysteic acid residues. Glu-C is a serine proteinases that cleaves with high specificity at the C-terminus of glutamic and aspartic acids. Use of alternative enzymes for protein digestion increases the confidence in mass spectrometry data by confirming the protein sequence and aids in the mapping of
post-translational modifications (PTMs). Here we describe the cleavage specificity of endoproteinases Asp-N and Glu-C, and show an example of improved protein sequence coverage through the use of alternative digestion with these enzymes.
Introduction
Endoproteinases Asp-N and Glu-C have been used for protein characterization for over 30 years and have gained importance recently due to advancements in mass spectrometry techniques. Asp-N preferentially cleaves proteins at the N-terminus of aspartic and cysteic acid (1–3). Glu-C cleaves at the C-terminus of glutamic and aspartic residues (4–6). Due to their specific cleavage sites, these proteinases create unique peptide fragments available for mass spectrometry analysis. Comparing protein sequences or mapping data from Asp-N or Glu-C digests to that of other proteinases promotes higher confidence in data (7–12).
Protein digestion is required for either the bottom-up or middle-down approach to protein analysis. In the bottom-up approach, the optimal peptide size range for analysis is 600–5,000Da. Recent advancements in mass spectrometry such as electron transfer dissociation (ETD) and electron capture dissociation (ECD), the potential to combine with collision activated dissociation (CAD) and in software, have expanded the peptide size range to 600–20,000Da. These advancements have promoted the middle-down approach to protein analysis (9, 13–16). However, some peptides still may fall above or below the desired peptide size range. This results in decreased protein coverage and incomplete data collection.
To increase protein coverage, additional proteinases have been used. Using an alternative proteinases individually or in combination with other proteinases creates a unique peptide map that may include sequences not seen in trypsin digestions. Overlaying peptides obtained from digestion with alternative proteinases increases protein coverage and overall confidence in protein identification. When alternative enzymes are used, larger peptides are cleaved into smaller fragments, which are more manageable for the instrumentation. Conversely, protein sections cleaved into peptides too small for analysis by one enzyme might be cleaved into larger peptides with a different enzyme. Alternative proteinases also help to overcome incomplete digestion caused by PTMs, which prevent the proteinases from accessing that particular site. By using alternative enzymes, the protein might be cleaved at sites further away from the PTM. The examples above show that alternative proteinases are beneficial to protein analysis (9, 13–16).
Asp-N, Sequencing Grade
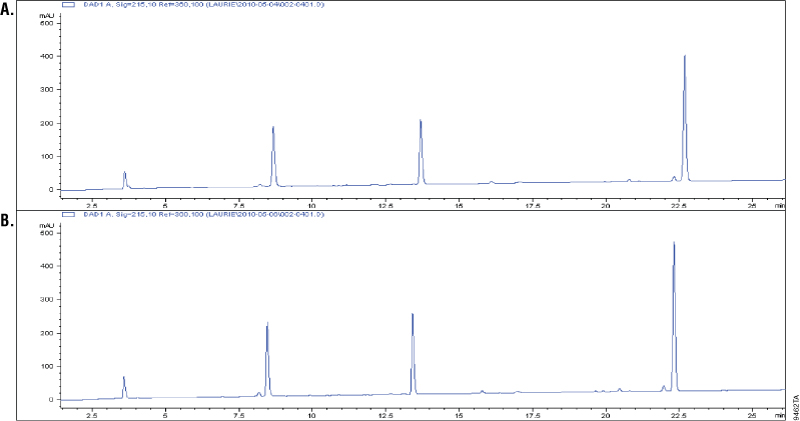
Asp-N is a metalloprotease purified from Pseudomonas fragi. Asp-N cleaves at the N-terminus of aspartic and cysteic acid residues with high specificity (1–3). Cysteic acid residues are an oxidized form of the cysteine residues Additional cleavage has been reported at glutamic residues; however, the rate of cleavage at aspartic acid is 2,000-fold higher than at the glutamyl residues (2–3). To demonstrate cleavage specificity, we over digested glucagon using a 1:10 Asp-N to peptide ratio (w/w). Samples from 1 and 18 hours of digestion were analyzed using C18 reverse-phose chromatography and compared. Digestion of glucagon with Asp-N creates four peptides. A change in the cleavage pattern would indicate nonspecific cleavage by Asp-N. The results showed the four peaks were unchanged after 18 hours of digestion, which indicated high cleavage specificity (Figure 1).
Glu-C, Sequencing Grade
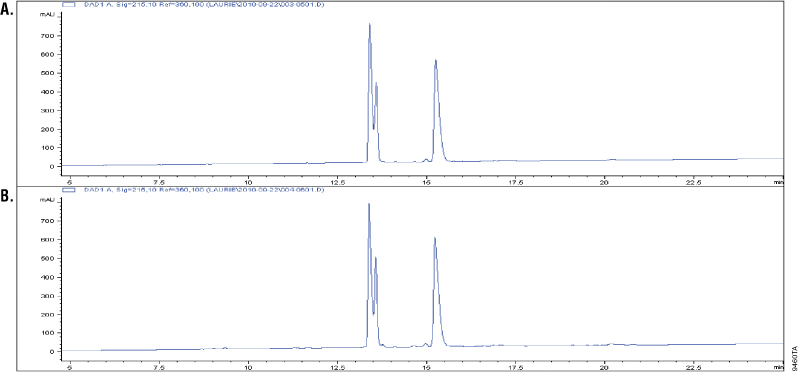
Glu-C is a serine proteinase purified from Staphylococcus aureus V8. It cleaves specifically at the C-terminus of glutamic and aspartic acids (4–6). Buffer composition effects the specificity of Glu-C. In phosphate buffers, both glutamic and aspartic residues are cleaved; however, in ammonium bicarbonate and ammonium acetate buffers (pH 4.0), only the glutamic residues are cleaved (6). The specificity of Glu-C was demonstrated by overdigesting insulin β-chain in a 1:10 Glu-C-to-protein ratio (w/w) in ammonium bicarbonate buffer. The digestion of insulin β-chain by Glu-C creates three peaks in the C18 reverse-phased analysis. Additional peaks in the chromatographs would indicate nonspecific cleavage activity. Figure 2 shows that the peak patterns apparent after 1 hour of digestion (Panel A) remain unchanged after 18 hours of digestion, indicating cleavage specificity (Panel B).
Multiple Enzyme Digestion
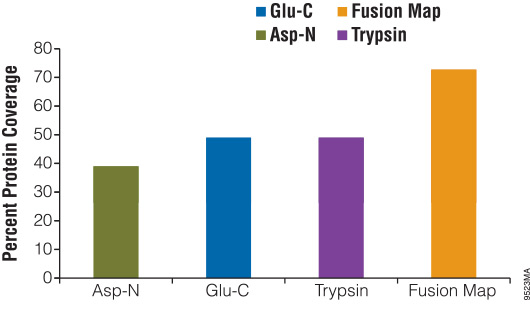
Digesting a protein with alternative enzymes, either individually or in combination, is useful for increasing the sequence coverage of a protein, identifying PTM sites and identifying translocation partners. To demonstrate the use of alternative proteinases for increasing protein sequence coverage, we digested β-casein with either Asp-N, Glu-C or trypsin for 18 hours. The reactions were stopped and analyzed using liquid chromatography/mass spectrometry/mass spectrometry (LC/MS/MS). The results of the digestions were as follows: Glu-C, 49%; Asp-N, 37%; and trypsin, 49%. After overlaying the peptide maps obtained from each digestion, the total coverage increased to 73% (Figure 3).
Conclusion
We show here that Asp-N and Glu-C, Sequencing Grade, endoproteinases (Cat. #V1621 and V1651) are excellent tools that can be used to improve protein analysis. Both proteinases demonstrate high cleavage specificity. Asp-N, Sequencing Grade, proteinase specifically cleaves at the N-terminus of the aspartic and cysteic acid residues. Glu-C, Sequencing Grade, proteinase cleaves at the C-terminus of glutamic and aspartic residues. These enzymes create different peptides compared to those created by trypsin digestion, offering higher protein coverage and an overall increase in protein identification. In the model experiment with β-casein, we showed that incorporation of Asp-N and Glu-C into the digestion protocol led to a 24% increase in protein coverage when compared with the coverage with trypsin alone. Asp-N and Glu-C, Sequencing Grade, endoproteinases also offer an efficient way to protein characterization, protein sequence coverage and PTM mapping.
References
- Drapeau, G.R. (1980) Substrate specificity of a proteolytic enzyme isolated from a mutant of Pseudomonase fragi. J. Biol. Chem. 255, 839–40.
- Ingrosso, D. et al. (1989) Specificity of endoproteinase Asp-N (Pseudomonas fragi): cleavage at glutamyl residues in two proteins. Biochem. Biophys. Res. Commun. 162, 1528–34.
- Geu, B.U. et al. (1990) Characterization of glutamyl cleavage activity of endoproteinase Asp N. J. Prot. Chem. 9, 299–300.
- Drapeau, G.R. (1972) Purification and properties of an extracellular protease of Staphylococcus aureus. J. Biol. Chem. 247, 6720–6.
- Drapeau, G.R. (1978) The primary structure of staphylococcal protease. Can. J. Biochem. 56, 534–44.
- Drapeau, G.R. (1977) Cleavage at glutamic acid with staphylococcal protease. Methods Enzymol. 47, 189–91.
- Fischer, F. and Poetsch, A. (2006) Protein cleavage strategies for an improved analysis of the membrane proteome. Proteome Sci. 4, 2–14.
- Wu, C.C. and MacCoss, M.J. (2002) Shotgun proteomics: Tools for the analysis of complex biological systems. Curr. Opin. Mol. Ther. 4, 242–50.
- Swaney, D.L., Wenger, C.D. and Coon, J.J. (2010) Value of using multiple proteases for larger-scale mass spectrometry-based proteomics. J. Proteome Res. 9, 1323–9.
- Choudhary, G. et al. (2003) Multiple enzymatic digestion for enhanced sequence coverage of proteins in complex proteomic mixtures using capillary LC with ion trap MS/MS. J. Proteome Res. 2, 59–67.
- Elenitoba-Johnson, K.S. et al. (2006) Proteomic identification of oncogenic chromosomal translocation partners encoding chimeric anaplastic lymphoma kinase fusion proteins. Proc. Natl. Acad. Sci. USA 103, 7402–7.
- Biringer, R.G. et al. (2006) Enhanced sequence coverage of proteins in human cerebrospinal fluid using multiple enzymatic digestion and linear ion trap LC-MS/MS. Brief Funct. Genomic. Proteomic. 5, 144–53.
- Young, N.L., Plazas-Mayorca, M.D. and Garcia, B.A. (2010) Systems-wide proteomic characterization of combinatorial post-translational modification patterns. Expert Rev. Proteomics 7, 79–92.
- Mann, M. and Kelleher, N.L. (2008) Precision proteomics: The case for high resolution and high mass accuracy. Proc. Natl. Acad. Sci. USA 105, 18132–8.
- Meyer, B. Papasotiriou, D.G. and Karas, M. (2010) 100% protein sequence coverage: A modern form of surrealism in proteomics. Amino Acids 41, 291–310.
- Cannon, J. et al. (2010) High-throughput middle-down analysis using an orbitrap. J. Prot. Res. 9, 3886–90.