Isolation of Genomic DNA from Agricultural Bacteria Using the Wizard® MagneSil™ Plasmid Purification System
Promega Corporation
Publication Date: 2001
Abstract
We have successfully adapted the Wizard® MagneSil™ Plasmid DNA Purification System (Cat.# A1635) to isolate genomic DNA from soil bacteria important in research and the agricultural industry. In this article, we describe the modified protocol for isolating genomic DNA from soil bacteria using manual and automated approaches on the Biomek® 2000. The isolated genomic DNA was then used to PCR amplify an 875bp DNA fragment.
Introduction
The Wizard® MagneSil™ DNA Purification System uses silica-coated paramagnetic particles to reversibly bind and purify DNA away from cell debris and proteins released upon directed cell lysis, alkaline denaturation and neutralization. MagneSil™ technology is currently used to isolate and purify plasmid DNA (in bacteria) and to purify DNA from sequencing reactions as well as following PCR.
Scientists working on agricultural studies, as well as ecologists, need to be able to routinely isolate genomic DNA from soil bacteria for gene structure studies as well as for taxonomical identification of bacterial species.
We have developed a simplified protocol for the isolation and purification of PCR quality genomic DNA from soil bacteria using Wizard® MagneSil™ RED (Cat.# A1641). Yields of genomic DNA ranged from 10-100+ng. The experiments here outline both the manual and automated procedures and demonstrate robust PCR product amplification from minimal volume amounts of final eluate.
Materials and Methods
Manual Protocol:
Genomic DNA was isolated from Acetobacter sp., Citrobacter freundi, Rhyzobium leguminosarum, Azotobacter venelandi, Rhodospirillum rubrum, Enterobacter sp., and E. coli strain JM109, using components of the Wizard® MagneSil™ Plasmid Purification System and other solutions. An 875bp DNA fragment corresponding to a region of the 16S ribosomal DNA sequence was amplified from the genomic DNA by PCR using region-conserved primers.
Experiment 1: Lyse cells directly using MagneSil™ RED solution.
Experiment 2: Treat cells first with Nuclei Lysis Solution (Cat.# A7943) containing 5µl of RNase A (Cat.# A7973). Then add MagneSil™ RED solution. Optional addition of RNase A may help increase yield of DNA, as RNA competes with DNA for binding to MagneSil™ paramagnetic particles.
- 10ml cultures of Acetobacter sp., Citrobacter freundi, Rhyzobium leguminosarum, Azotobacter venelandi, Rhodospirillum rubrum and Enterobacter sp. E. coli JM109 (as control) were grown initially from slants using a 2 day incubation at 25°C followed by a 1:100 dilution (noted 1:100) and re-grown overnight.
- Cultures were incubated with shaking in ATCC 111 Rhizobium X Medium or Nutrient broth with soil water medium (Carolina BA-15-3785).
- Centrifuge 1ml of each culture in a 1.5ml microcentrifuge tube. Use a deep well 96 well plate for many manual samples, e.g. Costar® 431191 Deep Well).
- Resuspend cell pellet in 120µl Cell Resuspension Solution (Cat.# A7114) containing 3mg/ml of lysozyme. Incubate 15 minutes at room temperature. For many manual samples, use 96 well U-bottom wash plate (Greiner 650101-96).
Protocol 1: Add 50µl MagneSil™ RED and vortex to mix. Incubate 10 minutes at room temperature. Total volume is now 170µl.
Protocol 2: Add 100µl of Nuclei Lysis Solution and 5µl RNase A, vortex briefly to mix and incubate 5 minutes at room temperature. Add 50µl MagneSil™ RED and vortex to mix. Incubate 10 minutes at room temperature. Total volume is 270µl.
Protocol 1 and 2: Capture the lysate on MagneSphere™ Technology Magnetic Separation Stand (Cat.# Z5332 or Z5342). Alternatively, use the MagnaBot® 96 Magnetic Separation Device (Cat.# V8151) for 96 well plate format.
Remove the supernatant. Wash three times with 150µl of 70% ethanol, removing supernatant each time.
Dry at room temperature for 10 minutes, or 65°C for 5 minutes.
Resuspend the captured DNA in 100µl Nuclease-Free Water (Cat.# P1193), capture and collect the resuspended DNA into a 1.5ml microcentrifuge tube (or 96 well plate).
Measure O.D.260 and O.D.280 (optional).
PCR Amplification: Perform PCR using the 16S rDNA-specific primers (515F and 1390R). Primers were chosen to amplify highly conserved sequences among most bacterial species (1) . 16S Oligonucleotide Primers:
515F Forward: 5´-GTGCCAGCAGCCGCGGTAA-3´
1390 Reverse: 5´-AGGCCCGGAACGTATTCAC-3´
UV Treatment for PCR, to inhibit amplification of background DNA contamination in PCR solutions:
Treat the following materials and reagents with UV light for 20 minutes:
- Master Mix without dNTPs
- Nuclease-Free Water (Cat.# P1193) with cap off
- GeneAmp® 0.5ml amplification tubes
- Pipettes
- 1000P, 200P and 20P pipette tips with aerosol barrier
- Gloves
- Tube racks
- Ice in containers
- Mineral oil (with cap off)
After UV treatment, add dNTPs to the master mix and assemble PCR.
| PCR Master Mix. (15 reactions for samples from each manual protocol, 1 and 2). | |
| Component | Volume |
| 10X Reaction Buffer | 75µl |
| MgCl2, 25mM | 75µl |
| PCR Mix dNTPs, 10mM each (Cat.# M1141) | 15µl |
| Taq DNA Polymerase | 3.3µl |
| Water to | 570µl |
PCR:
- 5µl genomic DNA sample, or nuclease-free water for no-template control reaction.
- 3.5µl 515F primer (13.8pmol/µl stock, final concentration 50pmol or water for no-primer control).
- 3.5µl 1390R primer (13.8pmol/µl stock, final concentration 50pmol or water for no-primer control).
- 7µl of nuclease-free water for no-primer control reaction.
- 38µl of PCR Master Mix.
- 50µl final volume.
- Samples were overlaid with a drop of mineral oil.
PCR Cycle:
- 2 minutes 94°C
- 30 cycles: 30 seconds 94°C, 1 minute 60°C, 1 minute 72°C
- 7 minutes 72°C
- Overnight 4°C
Automated Protocol
Biomek® 2000 Program for Isolation of Genomic DNA from Bacteria.
- Resuspend bacteria cell pellet in 120µl of Cell Resuspension Solution containing 3mg/ml lysozyme and incubate at room temperature for 10 minutes. (Note: Lysostaphin may be used at the same concentration for some gram-positive bacteria.)
- 100µl of Nuclei Lysis Solution is added to the resuspended bacterial cell pellet and incubated at room temperature for 2 minutes to allow for cell lysis.
- 50µl of MagneSil™ RED is then added to bacterial cell lysate and mixed to capture bacterial genomic DNA. The cell lysate containing the MagneSil™ RED is then transferred to the 96-well U-bottom wash plate (Greiner 650101-96). Once the lysate has been transferred to the wash plate, the plate is moved to the MagnaBot® and the MagneSil™ containing genomic DNA is separated from cell lysate. The cell lysate is removed to the waste reservoir.
- The MagneSil™ particles containing the captured genomic DNA are then washed three times with 70% ethanol using the Wash 8 tool that is connected to a bottle containing 25ml of 70% ethanol. After the third wash, the MagneSil™ particles are air-dried to remove residual ethanol.
- Finally, the genomic DNA is eluted in 100µl of nuclease-free water and placed into the 96-well elution plate.
 Figure 1. Initial deck configuration of the Biomek® 2000.
Figure 1. Initial deck configuration of the Biomek® 2000.
The tools required include the Wash 8, MP200 and Gripper all placed at position A1. In addition, a 6-port valve is required. Positions A5, A6 and B6 remain empty. A MagnaBot® 96 Magnetic Separation Device (Cat.# V8151) is placed at position A2. Boxes of P250 tips are placed at positions A3 and A4. Position B1 contains a reservoir for waste disposal. A 96-deep well plate containing pelleted bacterial culture with media removed is placed at position B2. At positions B3 and B5 are a 96 well U-bottom plate, one for the wash steps of the protocol and the other for purified sample elution. At position B4 are reservoirs containing the indicated volumes of reagents. *Add 3mg/ml lysozyme to the Cell Resuspension Solution (Cat.# A7114). **To the 4ml of Nuclei Lysis Solution (Cat.# A7941) add 133µl of RNase A (Cat.# A7973). Finally, a 25ml 70% ethanol bottle is attached to the 6-port valve.
Conclusions
We successfully isolated genomic DNA from soil bacteria using modified manual and automated protocols with the Wizard® MagneSil™ System. The DNA was used to amplify by PCR an 875bp fragment from the 16S bacterial ribosome DNA sequence. In both protocols, PCR amplification was robust across all samples after only 30 amplification cycles and using minimal amounts of genomic DNA product.
UV light treatment of tools and reagents used for setting up and performing PCR effectively prevented the background amplification of the 16S ribosomal DNA sequence from either the Taq DNA polymerase or from other non-specific contaminating bacteria. This is shown in Figure 2 (lane 4), in Figure 3 (lane 3), in Figure 4 (lane 3) and in Figure 5 (lane 3). PCR amplification was also performed with varying amounts of genomic DNA as demonstrated in Figure 5. Robust amplification was seen with either 1.0 O.D.600 units or 0.5 O.D.600 units of starting bacterial culture. [[For those samples with data for 0.5 O.D.600 units only, cultures did not grow to an O.D.600 of 1.0, thus not permitting those experiments.]] Use of Nuclei Lysis Solution and RNase A appeared to increase the PCR signal intensity (compare Figures 2 and 3). The automated protocol produced clean sharp bands in PCR.
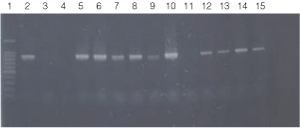 Figure 2. PCR amplification of bacterial genomic DNA from various soil bacteria, using manual protocol #1.
Figure 2. PCR amplification of bacterial genomic DNA from various soil bacteria, using manual protocol #1.
The PCR DNA fragment is 875bp.
Lane 1, 100bp DNA Ladder (Cat.# G2101);
lane 2, E. coli JM109 positive control
lane 3, blank;
lane 4, no-template negative control;
lane 5, Acetobacter sp. (tube #1);
lane 6, Citrobacter freundi (tube #2);
lane 7, Citrobacter freundi 1:100 (tube #3);
lane 8, Rhyzobium leguminosarum (tube #4);
lane 9, Rhyzobium leguminosarum 1:100 (tube #5)
lane 10, Azotobacter venelandi (tube #6);
lane 11, Azotobacter venelandi 1:100 (tube #7);
lane 12, Rhodospirillum rubrum (tube #8);
lane 13, Rhizobium leguminosarum (tube #9);
lane 14, Enterobacter sp. (tube #10);
lane 15, Citrobacter freundi (tube #11).
Bacteria identified in lanes 12-15 were grown in nutrient broth with soil water medium (Carolina BA-15-3785). Bacteria identified in all other lanes were grown in ATCC 111 Rhizobium X Medium.
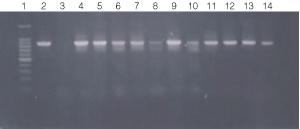 Figure 3. PCR amplification of bacterial genomic DNA from various soil bacteria, using manual protocol #2.
Figure 3. PCR amplification of bacterial genomic DNA from various soil bacteria, using manual protocol #2.
The PCR DNA fragment is 875bp.
Lane 1, 100 bp DNA ladder (Cat.# G2101);
lane 2, E. coli
JM109 positive control;
lane 3, No template negative control;
lane 4, Acetobacter sp.
(tube #1);
lane 5, Citrobacter freundi (tube #2);
lane 6, Citrobacter freundi
1:100 (tube #3);
lane 7, Rhyzobium leguminosarum (tube #4);
lane 8, Rhyzobium
leguminosarum 1:100 (tube #5);
lane 9, Azotobacter venelandi (tube #6);
lane
10, Azotobacter venelandi 1:100 (tube #7);
lane 11, Rhodospirillum rubrum
(tube # 8);
lane 12, Rhizobium leguminosarum (tube #9);
lane 13, Enterobacter
sp. (tube # 10);
lane 14, Citrobacter freundi (tube #11)
Bacteria identified in lanes 1-15 were grown in nutrient broth
with soil water medium (Carolina BA-15-3785).
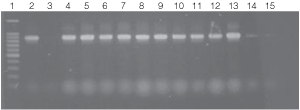 Figure 4. PCR amplifications of bacterial genomic DNA from various soil bacteria using the automated Biomek® 2000 protocol.
Figure 4. PCR amplifications of bacterial genomic DNA from various soil bacteria using the automated Biomek® 2000 protocol.
The PCR DNA fragment is 875bp.
Lane 1, 100bp DNA Ladder (Cat.# G2101);
lane 2, E. coli JM109 positive control DNA isolated using
manual protocol;
lane 3, negative control no template;
lanes 4 and 5, E. coli (Biomek® protocol);
lanes 6 and 7, Enterobacter sp.;
lanes 8 and 9, Azotobacter venelandi;
lanes 10 and 11, Rhyzobium leguminosarum;
lanes 12 and 13 Acetobacter;
lanes 14 and 15 Citrobacter freundi.
All bacteria were grown in ATCC 111 Rhizobium X
Medium.
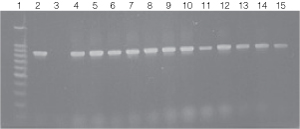 Figure 5. PCR amplification of bacterial genomic DNA from various soil bacteria using the automated Biomek® 2000 protocol.
Figure 5. PCR amplification of bacterial genomic DNA from various soil bacteria using the automated Biomek® 2000 protocol. The PCR DNA fragment is 875bp. Genomic DNA was
isolated from cultures grown to both 0.5 and 1.0 O.D.600 units.
Lane 1, 100bp DNA Ladder (Cat.# G2101);
lane 2, E. coli JM109 positive
control DNA isolated using the manual protocol;
lane 3, negative control no template;
lane
4, E. coli (Biomek®
protocol) O.D.600 (0.5);
lane 5, E. coli (Biomek® protocol) O.D.600 (1.0);
lane 6, Enterobacter O.D.600 (0.5);
lane 7, Enterobacter O.D.600 (1.0);
lanes 8 and 9, Azotobacter venelandi O.D.600
(0.5);
lane 10, Rhyzobium leguminosarum O.D.600 (0.5);
lane 11, Rhyzobium leguminosarum O.D.600 (1.0);
lanes 12 and 13, Acetobacter O.D.600 (0.5);
lanes 14 and 15, Citrobacter freundi O.D.600
(0.5).
All bacteria were grown in ATCC 111 Rhizobium X Medium.
Isolation of genomic DNA from soil bacteria is an important first step for agricultural scientists and ecologists involved in molecular biology research. We have adapted the Wizard® MagneSil™ DNA Purification System to permit isolation and purification of PCR-quality genomic DNA from soil bacteria. The adapted protocols provided here are quick, simple and flexible. Furthermore, the these protocols avoid organic solvents, which are messy as well as hazardous.
Related Products
Related Protocols
Related Resources
Article References
- Topp, E. et al. (2000) Characterization of an atrazine-degrading Pseudaminobacter sp. isolated from Canadian and French agricultural soils. Appl. Environ. Microbiol. 66, 2773–82.
How to Cite This Article
Scientific Style and Format, 7th edition, 2006
Farfan, A., Abramovici, H. and Grunst, T. Isolation of Genomic DNA from Agricultural Bacteria Using the Wizard® MagneSil™ Plasmid Purification System. [Internet] 2001. [cited: year, month, date]. Available from: https://www.promega.com/resources/pubhub/enotes/isolation-of-genomic-dna-from-agricultural-bacteria-using-the-wizard-magnesil-system/
American Medical Association, Manual of Style, 10th edition, 2007
Farfan, A., Abramovici, H. and Grunst, T. Isolation of Genomic DNA from Agricultural Bacteria Using the Wizard® MagneSil™ Plasmid Purification System. Promega Corporation Web site. https://www.promega.com/resources/pubhub/enotes/isolation-of-genomic-dna-from-agricultural-bacteria-using-the-wizard-magnesil-system/ Updated 2001. Accessed Month Day, Year.
Products may be covered by pending or issued patents. Please visit our patent and trademark web page for more information.