Minimizing PCR Cross-Contamination Using GoTaq® DNA Polymerase with dUTP and UNG
Robert Deyes
Promega Corporation
Publication Date: 2008
Abstract
A major source of PCR cross-contamination is carryover of PCR products in subsequent amplification reactions. One way to minimize such contamination is to include dUTP and uracil-DNA N-glycosylase in PCR. Here, we demonstrate this method with GoTaq® DNA Polymerase and suggest a protocol for routine laboratory use.
Introduction
PCR can detect trace amounts of target DNA, making it sensitive to DNA contamination, especially from previously amplified PCR products. A common method to minimize PCR cross-contamination uses dUTP and uracil-DNA N-glycosylase(a) (referred to as UNG or UDG) (1). This method involves two changes to basic PCR: dUTP is incorporated into the amplimer instead of dTTP, and the PCR mix is pretreated with UNG, which is subsequently heat-inactivated during PCR. UNG selectively degrades uracil-containing DNA, thus removing any PCR product contaminants and allowing amplification of only the target DNA.
GoTaq® DNA Polymerase is a proprietary formulation of Taq DNA polymerase that gives robust amplification; when used with the Green GoTaq® Reaction Buffer, you can go directly from PCR to gel analysis without adding sample buffer. This study tests the compatibility of the GoTaq® DNA Polymerase and Green GoTaq® Buffer with dUTP incorporation and UNG treatment.
Results
The β-actin target was successfully amplified in the first round of PCR by GoTaq® DNA Polymerase using all concentration ratios of dUTP:dTTP tested (Table 1 and Figure 1). When 200µM dUTP was used without any dTTP, the amplification was inconsistent (data not shown). We used the highest concentration of dUTP that consistently gave strong amplification (175µM dUTP + 25µM dTTP; Figure 1, Lane 5) for further testing.
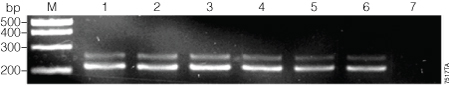
Figure 1. First-round amplification of the β-actin target with various ratios of dUTP to dTTP.
To assess the ability of GoTaq® DNA Polymerase to incorporate dUTP, reactions were set up with varying ratios of dUTP and dTTP at a final total concentration of 200 µM (Table 1). Thermal cycling conditions were: 35 cycles of denaturation at 95 °C for 30 seconds, annealing at 55 °C for 30 seconds and extension at 72 °C for 30 seconds. After PCR was completed, 5 µl of product was run on a 0.8% agarose gel. Lane M, PCR Markers (Cat.# G3161); lane 1, 0 µM dUTP:200 µM dTTP; lane 2, 100:100; lane 3, 125:75; lane 4, 150:50; lane 5, 175:25; lane 6, 200:0; lane 7, no‑template control.
| Reaction Components | Volume per 50 µl Reaction | |||||
| GoTaq® DNA Polymerase | 0.25 µl | |||||
| 5X Green GoTaq® Reaction Buffer | 10 µl | |||||
| 50 µM β‑Actin Primers | 1 µl | |||||
| dNTP Mix (dATP, dCTP, dGTP at 2 mM each) | 5 µl | |||||
| dUTP:dTTP (µM) | 0:200 | 100:100 | 125:75 | 150:50 | 175:25 | 200:0 |
| 2 mM dTTP | 5 µl | 2.5 µl | 1.9 µl | 1.2 µl | 0.7 µl | 0 µl |
| 2 mM dUTP | 0 µl | 2.5 µl | 3.1 µl | 3.8 µl | 4.3 µl | 5 µl |
| 2 ng/µl Human Genomic DNA (Cat.# G3041) | 1 µl | |||||
| Nuclease‑Free Water | to 50 µl | |||||
The conditions used in the second round of PCR ensured that no amplification would be seen if UNG successfully degraded the uracil‑containing amplimer. As shown in Figure 2, no PCR product was seen when the uracil‑containing product (175 µM dUTP + 25 µM dTTP) was treated with UNG and amplified (Figure 2, Lane 3). An amplimer resulted when UNG was left out of the reaction (Figure 2, Lane 2).
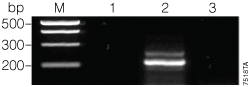
Figure 2. Second amplification of the 175 µM dUTP:25 µM dTTP PCR product.
To test the ability of UNG to degrade the uracil-containing GoTaq®-amplified PCR product in the Green GoTaq® Reaction Buffer, we performed a second round of amplification using UNG treatment and a trace amount of product from the first round of PCR. Uracil DNA Glycosylase (Invitrogen, Cat.# 18054-015) was used according to the manufacturer’s recommendations: PCR conditions were modified by an initial 10-minute step with 1 µl of UNG incubated at 37 °C and a final 72 °C soak after thermal cycling (see Figure 1). A volume of the first PCR was chosen to ensure the amount of genomic DNA template from the first reaction would be below the limit of detection for the conditions used in the second reaction (data not shown). The PCR was performed in a volume of 50 µl, and an aliquot of 5 µl was run on a 0.8% agarose gel. Lane M, PCR Markers (Cat.# G3161); lane 1, no-template control; lane 2, amplification without UNG treatment; lane 3, amplification with UNG treatment.
Conclusions
This study demonstrates that the GoTaq® DNA Polymerase and Green GoTaq® Reaction Buffer are compatible with both dUTP incorporation and subsequent UNG treatment. For consistent amplification success, trace amounts of dTTP should be included in the reaction mix with the dUTP (e.g., 25 µM dTTP + 175 µM dUTP; Figure 1). GoTaq® DNA Polymerase along with routine laboratory use of the dUTP incorporation and UNG treatment not only minimizes PCR product carryover but also provides researchers with the added benefit of directly loading PCR products onto an agarose gel.
References
- Longo, M.C., Berninger, M.S. and Hartley, J.L. (1990) Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene 93, 125–8.
Learn more about Taq DNA Polymerase and endpoint PCR.