Anti-HiBiT Magnetic Beads and DrkBiT Elution Peptide
For Convenient Affinity Purification of HiBiT-Tagged Proteins
- High-affinity enrichment of HiBiT-tagged proteins (Kd ≈ 6pM)
- Compatible with IP, co-IP and proteomics applications
- Gentle, native elution using DrkBiT peptide
- Validated for Western blotting, FACS and mass spectrometry
- Streamlined magnetic workflow: bind, wash and elution steps
Catalog Number:
Choose Magne® Beads or Elution Peptide
Size
Catalog Number: N7300
Catalog Number: N7301
Catalog Number: N7400
Efficient Enrichment of HiBiT-Tagged Proteins for IP, Co-IP and Proteomics
The HiBiT protein tag is a compact 11-amino acid peptide that binds with high affinity to LgBiT in the NanoBiT® system, reconstituting NanoLuc® Luciferase for highly sensitive luminescent detection. Its minimal size makes HiBiT ideal for CRISPR/Cas9 knock-ins, cloning vectors and synthetic constructs, enabling the expression and quantification of HiBiT-tagged proteins in native or engineered systems.
High-Affinity Capture with Anti-HiBiT Antibody and Magne® Beads
The Anti-HiBiT Monoclonal Antibody binds the HiBiT epitope with picomolar affinity (~6pM), ensuring strong and specific detection in immunoblotting, immunofluorescence, immunoprecipitation (IP) and flow cytometry (FACS).
When covalently linked to magnetic cellulose particles, it forms Anti-HiBiT Magne® Beads, which are a fast, convenient reagent for capturing and enriching HiBiT-tagged proteins from mammalian, yeast or bacterial lysates.
The magnetic format streamlines bind, wash and elution steps, making it ideal for IP, co-IP and downstream analyses such as Western blotting or mass spectrometry.
Study Protein:Protein Interactions with the HiBiT System
Anti-HiBiT Magne® Beads enable enrichment of HiBiT-tagged proteins together with their native binding partners, allowing co-immunoprecipitation (co-IP) and interactomics studies.
Combined with proteomic workflows such as immunoprecipitation-mass spectrometry (IP-MS) or affinity purification-mass spectrometry (AP-MS), this approach supports in-depth analysis of protein:protein interactions (PPI) and complex composition.
Flexible Elution Options for Downstream Applications
Several elution strategies are available:
- Low-pH buffer elution for efficient release prior to MS analysis
- SDS loading buffer for denaturing recovery
- DrkBiT Elution Peptide for gentle, competitive elution under native conditions
Using DrkBiT preserves protein structure and activity, supporting downstream assays such as enzymatic studies or structural analysis. The elution method can be tailored to experimental needs, with peptide elution for activity retention or acidic elution for proteomics workflows.
Together, Anti-HiBiT Magne® Beads and Elution Peptide provide a flexible, high-affinity system for enriching HiBiT-tagged proteins, supporting workflows that range from standard IP to advanced proteomics and interactomics research.
A.
B.

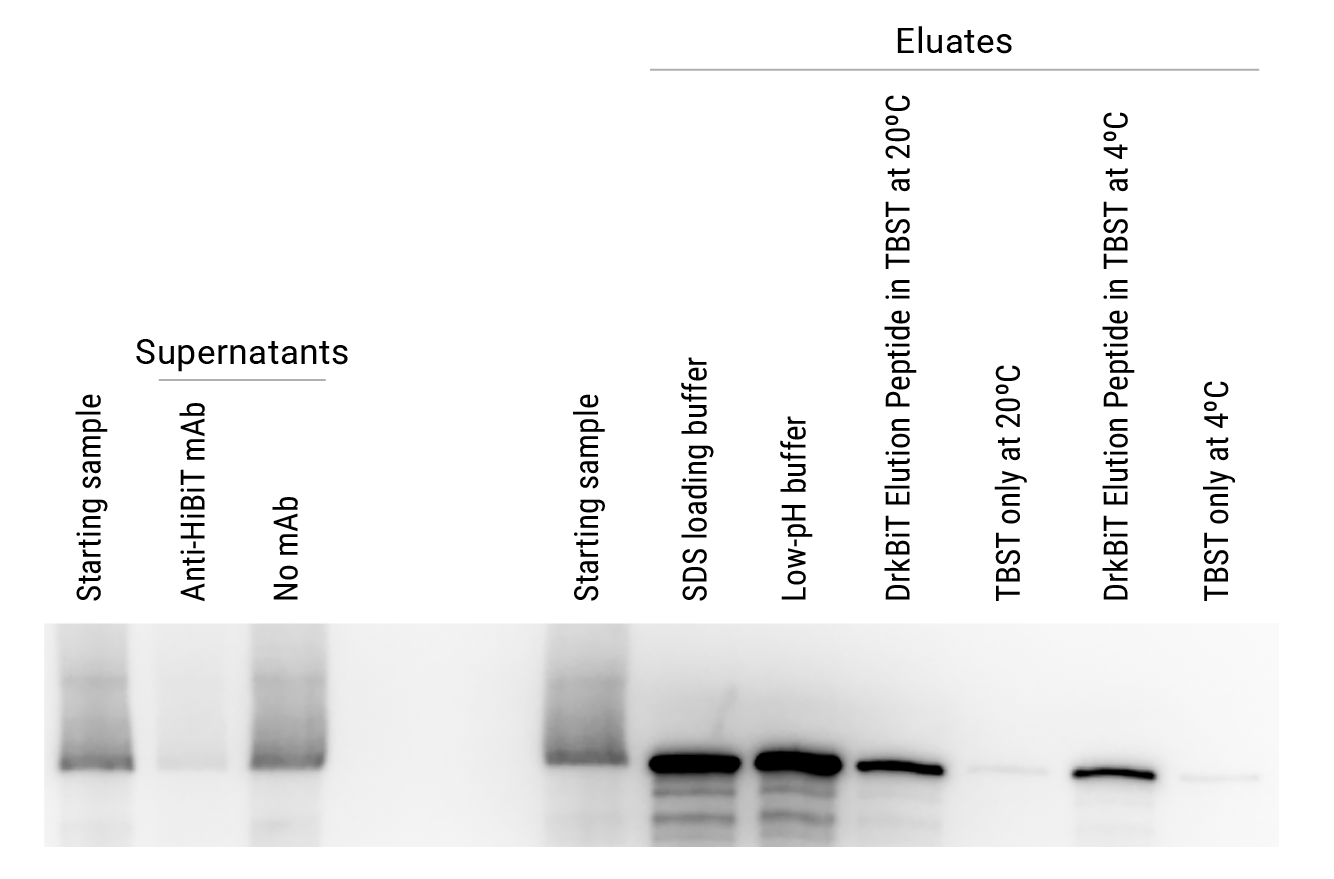
Comparison of elution strategies for HiBiT-tagged firefly luciferase immunoprecipitated from HEK293 cells. Starting samples consisted of lysate containing 5 × 106 cells/ml in Mammalian Lysis Buffer (Cat.# G9381) supplemented with Protease Inhibitor Cocktail (Cat.# G6521) and RQ1 RNase-Free DNase (Cat.# M6101). Cleared lysate was incubated with beads for 2 hours at 4°C. Eluates were generated using four different elution conditions: 1) SDS loading buffer; 2) low-pH buffer; 3) DrkBiT Elution Peptide (Cat.# N7400, diluted 100-fold in TBST) incubated for 2 hours at room temperature; or 4) DrkBiT Elution Peptide incubated overnight at 4°C. TBST-only buffer controls were performed for comparison to DrkBiT Peptide eluates. Eluates were generated using a 4-fold lower volume (100µl) than the starting lysate samples (400µl). Equivalent volumes of the starting lysate sample, supernatants and eluates were used for analysis. Panel A shows analysis of samples using the Nano-Glo® HiBiT Blotting System (Cat.# N2410). The left gel photo is scaled to lower intensities to illustrate effective clearance of Fluc-HiBiT from supernatants by Anti-HiBiT Magne® Beads but not control beads without antibody. The right gel photo is scaled to higher intensities to demonstrate increased concentrations of Fluc-HiBiT in the eluates compared to the starting sample. In Panel B, the enzymatic activity of firefly luciferase was measured using the ONE-Glo™ Assay (Cat.# E6110), highlighting the ability to gently elute HiBiT-tagged proteins and maintain enzyme activity using the DrkiBiT Elution Peptide.
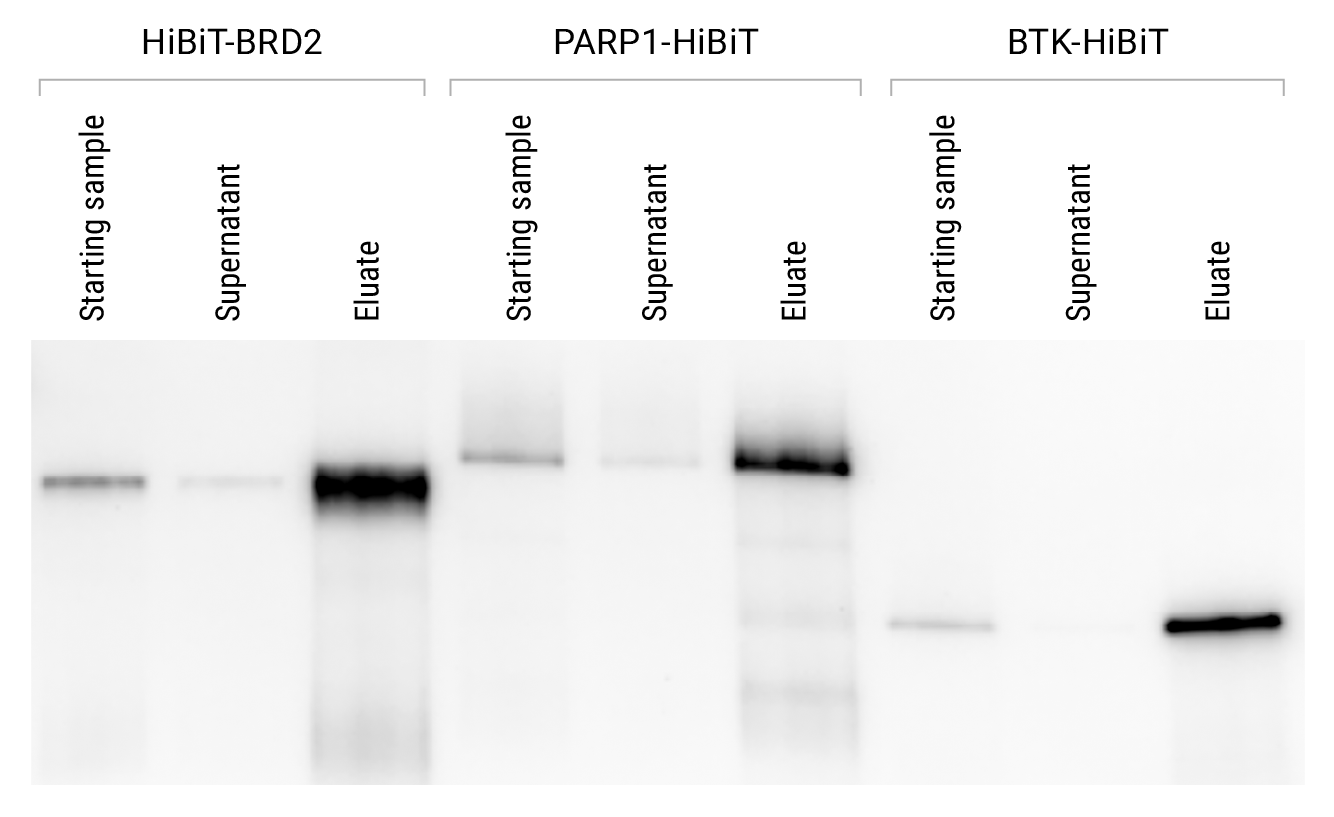
CRISPR-mediated HiBiT-tagging enables immunoprecipitation of proteins at endogenous levels. Using CRISPR/Cas9 genome-editing technology, the HiBiT tag was added to termini of BRD2, PARP1 and BTK in HEK293, DLD1 and K562 cells, respectively. The standard protocol in Section 3.B of Technical Manual #TM771 was used to immunoprecipitate the proteins. Samples were analyzed to monitor protein clearance from the supernatant and protein recovery in the eluate. Starting samples of cleared cell lysate (1ml; 5 × 106 cells/ml) were immunoprecipitated by binding overnight at 4°C and eluting in 80µl of 0.1M glycine (pH 2.5). Eluates were removed from beads and neutralized with 20µl of 2M Tris (pH 7.5). Equivalent volumes of each starting lysate sample, supernatant and eluate were analyzed using the Nano-Glo® HiBiT Blotting System (Cat.# N2410).

HiBiT-based IP-MS identifies SMARCA2-associated SWI/SNF proteins. The HiBiT tag was inserted at the C-terminus of endogenous SMARCA2 in HeLa cells using CRISPR/Cas9 genome editing. Triplicate immunoprecipitations were performed from lysates of both edited and parental cells using the standard protocol described in Section 3.B of Technical Manual #TM771. Eluates were processed with the Magnetic Proteomics Sample Prep Kit (Cat.# CS3325A04) and digested with Trypsin/Lys-C (Cat.# V5071). Peptides were analyzed by LC-MS/MS on a Thermo Fisher Scientific Orbitrap Exploris 240. Data were searched using ProteomeDiscoverer 3.1 against the Uniprot Human database using the Sequest search algorithm. Red circles in the volcano plot highlight known SMARCA2 interactors, including members of the SWI/SNF chromatin remodeling complex.
Simplify Your Workflow
Explore premade solutions to simplify your workflow—whether you need ready-to-use HiBiT-tagged protein vectors or prebuilt knock-in CRISPR cell lines designed for reliable expression and detection.
Frequently Asked Questions
Q: What are Anti-HiBiT Magne® Beads used for?
A: Anti-HiBiT Magne® Beads are designed to capture, enrich and purify HiBiT-tagged proteins from cell lysates for immunoprecipitation (IP), co-Immunoprecipitation (co-IP), Western blotting and mass spectrometry (IP-MS).
Q: How do Anti-HiBiT Magne® Beads work?
A: Anti-HiBiT Magne® Beads are coated with an antibody that binds specifically to the HiBiT epitope tag. When mixed with cell lysates, the beads pull down HiBiT-tagged proteins, which can then be washed and analyzed.
Q: What are my options for eluting proteins from Anti-HiBiT Magne® Beads?
A: The options for eluting proteins from Anti-HiBiT Magne® Beads are listed below:
• Low-pH buffer: robust release with minimal antibody contamination
• SDS loading buffer: denaturing elution, may co-elute antibody (especially with reducing agents, DTT, b-ME, TCEP)
• DrkBiT Elution Peptide: gentle, competitive release under native conditions, best for preserving protein function and complexes
Q: Why is my HiBiT-tagged protein not clearing from the supernatant?
A: Possible causes include exceeded bead capacity, suboptimal lysis conditions, insufficient binding time or HiBiT tag inaccessibility within a protein complex. Try increasing bead volume, optimizing buffer conditions or extending incubation.
Q: Why is the amount of eluted protein lower than expected?
A: Low eluted protein amounts can result from protein degradation, harsh wash conditions or incomplete elution. Use protease inhibitors, keep samples cold, optimize buffer composition and consider a second elution step with SDS buffer.
Q: How can I avoid antibody contamination in my eluate?
A: Avoid harsh denaturing elution if downstream analysis is sensitive to antibody fragments. Instead, use DrkBiT Elution Peptide or a low-pH elution, which will release HiBiT-tagged proteins while minimizing antibody carryover.
Q: Why is an expected binding partner missing in my co-IP experiment?
A: Protein complexes may dissociate during binding or washing, or they may be present only in small fractions of cells. Optimize buffers (e.g., add glycerol), shorten incubation, keep samples at 4°C or overexpress the binding partner.
Q: How can I reduce nonspecific proteins in my eluate?
A: Increase wash stringency (salt, detergents or wash volume), and consider elution with DrkBiT Elution Peptide or low-pH buffers for greater specificity.
Q: Why use the DrkBiT Elution Peptide?
A: The DrkBiT Elution Peptide competes with the HiBiT-tagged protein for antibody binding, allowing proteins to be released under native, non-denaturing conditions. This is ideal for maintaining protein activity and interactions.
Q: Can the Anti-HiBiT Magne® Beads be reused?
A: Anti-HiBiT Magne® Beads are intended for single-use experiments to ensure maximum specificity and performance. Reuse is not recommended.
Protocols
Specifications
Catalog Number:
Contenido
| Item | Part # | Presentación |
|---|---|---|
Anti-HiBiT Magne® Beads |
N730A | 1 × 1ml |
SDS
Search for SDSCertificado de Análisis
Use Restrictions
For Research Use Only. Not for Use in Diagnostic Procedures.Condiciones de Almacenaje
Contenido
| Item | Part # | Presentación |
|---|---|---|
Anti-HiBiT Magne® Beads |
N730B | 1 × 5ml |
SDS
Search for SDSCertificado de Análisis
Use Restrictions
For Research Use Only. Not for Use in Diagnostic Procedures.Condiciones de Almacenaje
Contenido
| Item | Part # | Presentación |
|---|---|---|
100X DrkBiT Elution Peptide |
N740A | 1 × 100μl |
SDS
Search for SDSCertificado de Análisis
Use Restrictions
For Research Use Only. Not for Use in Diagnostic Procedures.Condiciones de Almacenaje
Resources
Featured Resource
ASMS 2025 Poster: Quantitative Profiling of Endogenous Protein Interactions Using HiBiT Tag and IP-MS
To extend HiBiT’s utility beyond quantification, we developed a high-affinity Anti-HiBiT Monoclonal Antibody and Anti-HiBiT Magne® Beads for immunoprecipitation and mass spectrometry (IP-MS). This integrated workflow allows quantitative capture of HiBiT-tagged proteins and their interactors, enabling detailed mapping of endogenous protein complexes, post-translational modifications, and signaling network dynamics. Here, we demonstrate how combining bioluminescent quantification with antibody-based enrichment facilitates robust, reproducible analysis of protein:protein interactions at physiological levels.

Related Products
Productos Similares
Anti-HiBiT Monoclonal Antibody
Anticuerpo monoclonal específico y altamente sensible que permite métodos basados en anticuerpos para la detección de la tag HiBiT.
N7200, N7210
Anti-HiBiT Monoclonal Antibody, XFD Fluorophore Conjugates
Efficient flow cytometry analysis of HiBiT-tagged proteins.
CS3278A02, CS3278A06
Nano-Glo® HiBiT Blotting System
Método luminiscente para la detección rápida de proteínas marcadas con HiBiT en blots, sin necesidad de anticuerpos.
N2410
Usado con frecuencia junto con
Nano-Glo® HiBiT Lytic Detection System
Método bioluminiscente para detectar la cantidad total de proteínas marcadas con HiBiT en la célula.
N3030, N3040, N3050
Magne® Protein G and Magne® Protein A Beads
Perlas magnéticas de afinidad con proteína A o G para la purificación de anticuerpos a partir de medios de cultivo, ascitis y muestras de suero.
G7471, G7472, G7473, G8781, G8782, G8783





