Sample Preparation Method for Accurate Analysis of Nonenzymatic PTMs in Biotherapeutic Proteins
Global Proteomics Product Manager, Promega Corporation
Publication Date: August 2017, tpub_186
Abstract
Biotherapeutic proteins accumulate nonenzymatic posttranslational modifications (PTMs) during manufacturing and storage. These PTMs include deamidation, disulfide bond scrambling and oxidation. Peptide mapping is commonly used to monitor these modifications. However, the sample preparation steps involved in peptide mapping are also sources of nonenzymatic PTMs. We developed the AccuMAP® Low pH Protein Digestion Kit for protein sample preparation under low pH conditions and demonstrate here how digestion under these conditions suppresses nonenzymatic PTMs.
Introduction
Nonenzymatic posttranslational modifications (PTMs) can spontaneously occur in biotherapeutic proteins during manufacturing and storage, affecting efficacy and stability of these proteins. The major nonenzymatic PTMs are deamidation, disulfide bond scrambling and oxidation (Figure 1).
 Figure 1. Nonenzymatic PTMs. Schematic illustrating deamidation, disulfide bond scrambling and oxidation, all of which can be introduced during peptide mapping.
Figure 1. Nonenzymatic PTMs. Schematic illustrating deamidation, disulfide bond scrambling and oxidation, all of which can be introduced during peptide mapping. Figure 2. Peptide mapping of antibody. Peptide mapping utilizes trypsin or alternative proteases to digest an antibody into smaller fragments (peptides). By analyzing these peptides with LC/MS-MS or UV HPLC, amino acid sequence, glycosylation and nonenzymatic PTMs are determined.
Figure 2. Peptide mapping of antibody. Peptide mapping utilizes trypsin or alternative proteases to digest an antibody into smaller fragments (peptides). By analyzing these peptides with LC/MS-MS or UV HPLC, amino acid sequence, glycosylation and nonenzymatic PTMs are determined.Artificial PTMs compromise the analysis of authentic nonenzymatic PTMs. The problem can be resolved by shifting the pH of the sample preparation steps into the acidic range. The chemical agents and proteases involved in the sample preparation process, however require alkaline pH.
Development of a kit that provided reproducible reduction, alkylation and digestion under low pH conditions required optimization of reaction conditions as well as the selection of corresponding reagents. Details relating to the development process are described here.
 Figure 3. Schematic diagram of sample preparation by digestion performed with the AccuMAP™ Low pH Protein Digestion Kit.
Figure 3. Schematic diagram of sample preparation by digestion performed with the AccuMAP™ Low pH Protein Digestion Kit. Reduction and Alkylation at Low pH
Common reducing and alkylating agents favor alkaline pH. Because alkaline pH induces deamidation and disulfide bond scrambling, we modified the reduction and alkylation procedure to be compatible with low pH. With the AccuMAP® Kit, reduction and alkylation steps are performed at pH 5.6–5.8. To ensure efficient reduction at low pH we used TCEP (Tris(2-carboxyethyl)phosphine), which maintains high reducing activity at low pH. Alkylation is performed with IAM (iodoacetamide). Activity of this reagent is decreased at low pH. To compensate for decreased IAM activity at low pH, alkylation is allowed to proceed through the digestion reaction.
Protein Digestion at Low pH
Trypsin and other proteases commonly used in peptide mapping sample preparation favor alkaline pH in order to efficiently digest proteins. To avoid artificial nonenzymatic PTMs induced at these conditions, we developed protocols for trypsin digestion at low pH. (With the AccuMAP® Kit pH is shifted to 5.2–5.4 at the digestion step.) Our studies show that low pH primarily inhibits tryptic cleavages at lysine sites whereas arginine cleavage sites are still efficiently digested. To restore trypsin cleavage efficiency at lysine sites we supplemented trypsin with a special, low pH resistant recombinant Lys-C (rLys-C) protease. By supplementing trypsin with low pH resistant rLys-C we achieved efficient tryptic digestion at low pH (Figure 4).
 Figure 4. Efficient digestion with AccuMAP™ Low pH Protein Digestion Kit. Rituximab was digested according to a conventional protocol (at pH 8) or with the AccuMAP™ Low pH Protein Digestion Kit and analyzed with LC/MS. The data show that digestion at low pH was comparable to digestion at conventional conditions.
Figure 4. Efficient digestion with AccuMAP™ Low pH Protein Digestion Kit. Rituximab was digested according to a conventional protocol (at pH 8) or with the AccuMAP™ Low pH Protein Digestion Kit and analyzed with LC/MS. The data show that digestion at low pH was comparable to digestion at conventional conditions.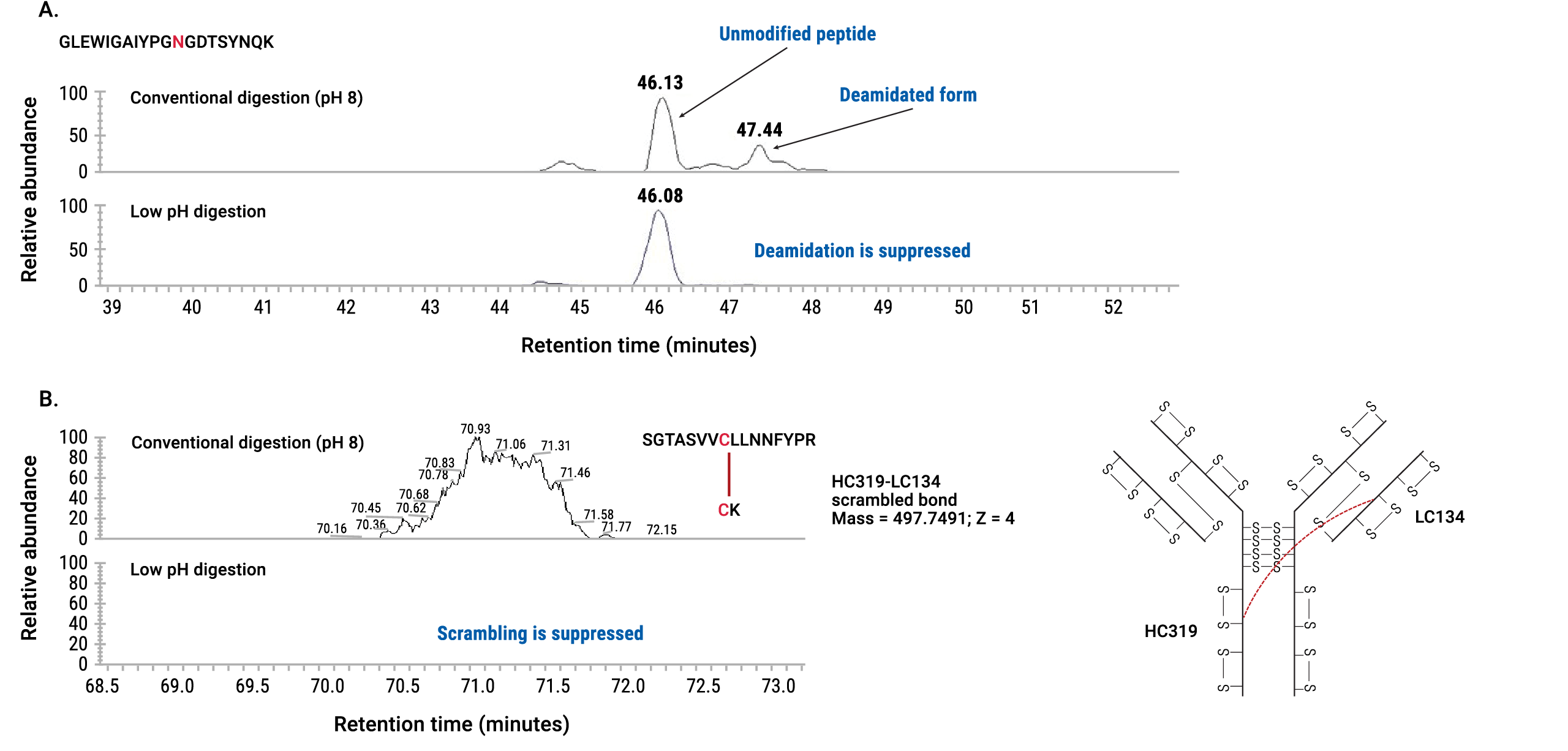 Figure 5. Suppression of deamidation and disulfide bond scrambling in IgG digested with AccuMAP™ Low pH Protein Digestion Kit. Panel A shows an extracted ion chromatogram of a GLEWIGAIYPGNGDTSYNQK peptide from rituximab digested at conventional conditions (pH 8) and at low pH with AccuMAP™ Low pH Protein Digestion Kit. The data show that asparagine (highlighted in red) was deamidated in this peptide at pH 8. In contrast, deamidation was fully suppressed at low pH. Panel B shows an extracted ion chromatogram of a peptide with a scrambled disulfide bond from panitumumab digested under conventional conditions (pH 8) and at low pH with the AccuMAP™ Low pH Protein Digestion Kit. Disulfide bond scrambling is evident at pH 8 while it is fully suppressed at low pH.
Figure 5. Suppression of deamidation and disulfide bond scrambling in IgG digested with AccuMAP™ Low pH Protein Digestion Kit. Panel A shows an extracted ion chromatogram of a GLEWIGAIYPGNGDTSYNQK peptide from rituximab digested at conventional conditions (pH 8) and at low pH with AccuMAP™ Low pH Protein Digestion Kit. The data show that asparagine (highlighted in red) was deamidated in this peptide at pH 8. In contrast, deamidation was fully suppressed at low pH. Panel B shows an extracted ion chromatogram of a peptide with a scrambled disulfide bond from panitumumab digested under conventional conditions (pH 8) and at low pH with the AccuMAP™ Low pH Protein Digestion Kit. Disulfide bond scrambling is evident at pH 8 while it is fully suppressed at low pH.Predigestion Step
Often, certain protein regions are resistant to proteolysis even after reducing disulfide bonds. This resistance is caused by the tightly folded conformation of these regions. Tight folding prevents protease access to internal cleavage sites. A common approach to overcome proteolytic resistance of tightly folded proteins or protein domains is predigestion with Lys-C protease under denaturing conditions. Digestion under denaturing conditions is possible due to the unique ability of Lys-C proteases to tolerate denaturing conditions. We used this tolerance to denaturing conditions to ensure efficient digestion of proteolytically resistant domains. As with all other steps in this protocol, predigestion is performed at low pH. Subsequently a denaturing agent is diluted and digestion is completed with trypsin or, optionally, rLys-C alone.
Reduction of Baseline Noise in Tryptic Digests
Baseline noise is a common problem in tryptic digests. The major cause of baseline noise is trypsin overdigestion, which results in the generation of semi-tryptic peptides. Semi-tryptic peptides are the products of specific cleavage at lysine or arginine residue at one terminus and nonspecific cleavage (primarily at an aromatic residue) at the other terminus.
Semi-tryptic peptides were evident in overnight trypsin digestions at 1:20 or lower trypsin:protein ratio in our study. We offer two potential solutions to this problem. One solution is to remove trypsin from a reaction and digest a protein with the AccuMAP® Low pH Resistant rLys-C alone. In contrast to trypsin, AccuMAP® Low pH Resistant rLys-C does not generate semi-tryptic peptides.
An alternative solution is based on optimization of the trypsin:protein ratio and digestion time. Optimal digestion was achieved at a 1:5 trypsin:protein ratio and 3-hour incubation. Baseline noise was minimal and digestion was efficient at these conditions (Figure 6).
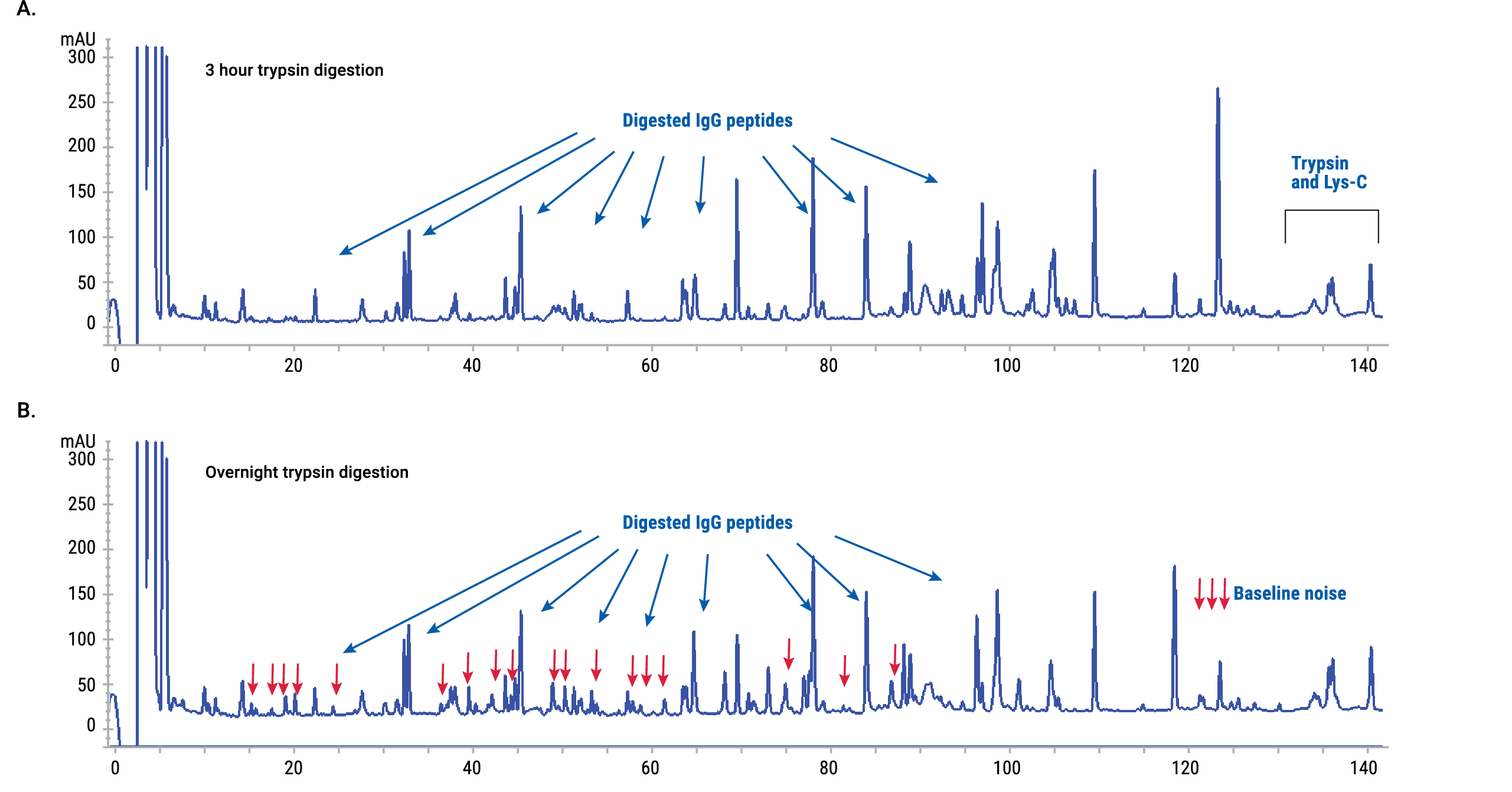 Figure 6. UV-HPLC chromatograms of Panitumumab digests. Panitumumab was predigested with AccuMAP™ Low pH Resistant rLys-C, then the reaction was diluted and digestion was completed by incubation with AccuMAP™ Modified Trypsin for 3 hours (Panel A) or overnight (Panel B). In this experiment we used a 1:5 trypsin:protein ratio. Note the accumulation of baseline noise for the overnight digest shown in Panel B.
Figure 6. UV-HPLC chromatograms of Panitumumab digests. Panitumumab was predigested with AccuMAP™ Low pH Resistant rLys-C, then the reaction was diluted and digestion was completed by incubation with AccuMAP™ Modified Trypsin for 3 hours (Panel A) or overnight (Panel B). In this experiment we used a 1:5 trypsin:protein ratio. Note the accumulation of baseline noise for the overnight digest shown in Panel B. Oxidation Suppression
Certain excipients and impurities have protein oxidation activity. For example, polysorbates, which are popular surfactants used to improve solubility of biotherapeutic proteins, decompose upon storage into hydrogen peroxide, which oxidizes proteins. This oxidation increases during proteolytic digestion due to the increased temperature. To suppress oxidation we recommend adding the optional AccuMAP® Oxidation Suppressant (included in the kit) to a reaction.
Conclusion
Nonenzymatic PTMs can be introduced during protein sample preparation for peptide mapping under conventional conditions. The AccuMAP® Low pH Protein Digestion Kit is designed to suppress artificial nonenzymatic PTMs. The entire procedure (i.e., reduction, alkylation and digestion) has been optimized for use at low pH. The digestion procedure is based on the use of trypsin and Low pH Resistant rLys-C. The combination of these two proteases provides maximum sequence coverage while minimizing baseline noise.
Related Products
Related Protocols
Related Resources
How to Cite This Article
Scientific Style and Format, 7th edition, 2006
Kobs, G. Sample Preparation Method for Accurate Analysis of Nonenzymatic PTMs in Biotherapeutic Proteins. [Internet] August 2017, tpub_186. [cited: year, month, date]. Available from: https://www.promega.com/es-es/resources/pubhub/sample-preparation-method-for-accurate-analysis-of-nonenzymatic-ptms-in-biotherapeutic-proteins/
American Medical Association, Manual of Style, 10th edition, 2007
Kobs, G. Sample Preparation Method for Accurate Analysis of Nonenzymatic PTMs in Biotherapeutic Proteins. Promega Corporation Web site. https://www.promega.com/es-es/resources/pubhub/sample-preparation-method-for-accurate-analysis-of-nonenzymatic-ptms-in-biotherapeutic-proteins/ Updated August 2017, tpub_186. Accessed Month Day, Year.