Improved Primer Pair for the SE33 Locus in the PowerPlex® ESI 17 Pro System
1Promega Corporation 2Human Identity Project Team, National Institutes of Standards and Technology
Publication Date: 2012
Introduction
In September 2009 Promega launched the PowerPlex® ESX and ESI Systems to meet the needs of the next-generation European STR genotyping systems (1) . These systems include five new loci (D1S1656, D2S441, D10S1248, D12S391 and D22S1045) recommended by the European Network of Forensic Science Institutes (ENFSI) and European DNA Profiling Group (EDNAP) for expanding the European Standard Set (ESS) loci (2) (3) . Both systems are available with or without primers to amplify the SE33 locus. The PowerPlex® ESX Systems are configured with the new ENFSI/EDNAP loci D10S1248, D22S1045 and D2S441 as miniSTRs (<125 bases) and D12S391 and D1S1656 as midiSTRs (125–185 bases) (4) . The PowerPlex® ESI Systems are configured with the other ESS loci as small as feasible while providing the new ENFSI/EDNAP loci as larger amplicons (5) .
Initial population studies performed in collaboration with the National Institutes of Standards and Technology (NIST) Forensic/Human Identity Project Team did not reveal any significant discordance between the PowerPlex® ESX and ESI Systems or with shared loci in other commercial kits (6) . Since this initial publication, there have been several reports of X.1 or X.3 alleles at the SE33 locus (where X represents an integer) instead of X or X.2, respectively, when using the PowerPlex® ESI 17 System (7) . In contrast, when these same DNA samples were genotyped with the PowerPlex® ESX 17 System, the correct allele call was made. To alleviate this discordance we developed a new SE33 primer set for inclusion in the PowerPlex® ESI 17 Pro System (Cat.# DC7780, DC7781).
X.1 and X.3 Single Nucleotide Polymorphisms in the SE33 Locus
During the initial concordance study performed with the PowerPlex® ESX and ESI Systems, none of the 1,455 samples used for concordance testing revealed any examples of a migration shift in an SE33 allele that would result in an X allele being called X.1 or an X.2 allele being called X.3 (6) . Since then, there have been several reports of a discordance when using the PowerPlex® ESI 17 System compared to the PowerPlex® ESX 17 System, PowerPlex® ES System, AmpFlSTR® SEfiler Plus™ Kit and AmpFlSTR® NGM SElect™ Kit (7) . The PowerPlex® ES System and AmpFlSTR® SEfiler Plus™Kit both use the Polymeropoulos primers (8) , which are the basis of most of the existing SE33 profiles generated in countries using this locus. This mutation appeared to be prevalent in individuals of West African origin, such as the US African-American population. Repeat genotyping of the original concordance study samples revealed nine samples of the original 448 US African-American samples that showed this type of mutation. These samples were improperly genotyped in the original study due to poor resolution on the capillary electrophoresis instrument. Analysis of another 46 US African-American samples revealed a further three discordant samples, for a total incidence of 2.43% in this population group. Analysis of GT37190 DNA (one of the original 448 US African-American samples obtained from NIST) with PowerPlex® ESX 17 and PowerPlex® ESI 17 Systems showed the migration difference that was observed (Figure 1).
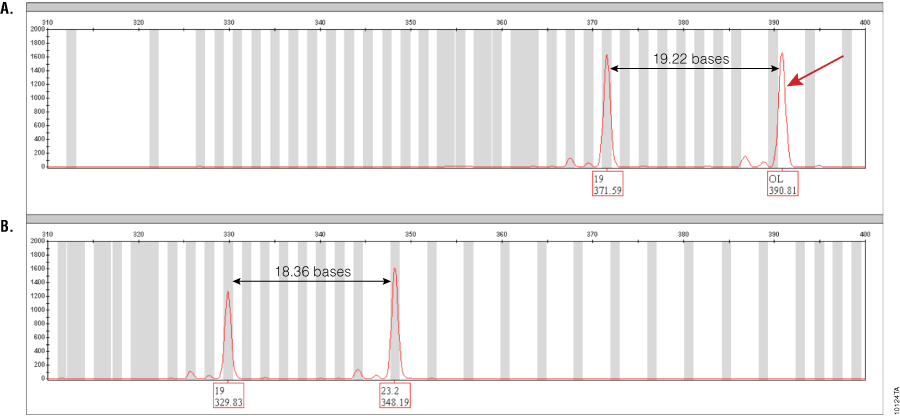 Figure 1. Amplification of GT37190 DNA with PowerPlex® ESI 17 and ESX 17 Systems.
Figure 1. Amplification of GT37190 DNA with PowerPlex® ESI 17 and ESX 17 Systems. The red dye channel encompassing the SE33 locus range for both PowerPlex® ESI 17 (Panel A) and ESX 17 (Panel B) amplifications of DNA sample GT37190 are shown. The 23.2 allele that runs off-ladder in PowerPlex® ESI 17 is indicated by a red arrow. Distances between alleles in both multiplexes are indicated. Peak labels show the allele call (top) and size in bases (bottom). The off-ladder peak seen with PowerPlex® ESI 17 is 0.94 bases larger than the corresponding position in the allelic ladder and hence falls outside of ±0.5-base binning window. This allele is correctly called as 23.2 with the PowerPlex® ESX 17 System.
The size of the affected allele ranged from 0.59 to 0.94 bases larger than the corresponding allele in the PowerPlex® ESI 17 Allelic Ladder, resulting in either an off-ladder allele call or an X.3 allele call if a virtual bin existed for that position in the ladder. The alleles affected in the twelve DNA samples mentioned were 12.2, 13.2, 15.2, 16.2, 23.2 and 25.2. We obtained two additional DNA samples from laboratories in Europe and identified a G to A transition located 68 bases downstream of the repeat (Figure 2). This matched the mutation found in 11 of the 12 samples identified at NIST. Other mutations that exerted the same effect are 60 bases (C to T) and 61 bases (G to A) downstream of the repeat. A closer analysis of the DNA sequence near these single nucleotide polymorphisms (SNPs) reveals the potential for a stem-loop structure, which would be destabilized by any of the three mutations identified. The existence of this stem-loop structure was demonstrated recently (9) . It is possible that this potential stem-loop structure and the predicted effect of the identified mutations could be responsible for the observed shift in migration, given the known effect of secondary structure on migration during capillary electrophoresis (10) .
 Figure 2. SE33 locus with the location of X.3 SNPs.
Figure 2. SE33 locus with the location of X.3 SNPs. The SE33 locus is shown with the location of the Polymeropoulos primers indicated in red. These are found in both the PowerPlex® ES System and AmpFlSTR® SEfiler Plus™ Kit. The repeat structure is in black and bold with the underlined portion representing the region used for determining allele number (sequence shown here is a 25.2 allele). Underlined sequences in red are bases whose mutation (G to A or C to T) at that position results in a shift in migration on the capillary relative to the allelic ladder of almost +1 base. Sequence in bold in larger font with inverted arrows represents a region of self-complementarity that is capable of forming a stem-loop structure in single-stranded DNA with a 5-base-pair stem and a 3-base loop.
The PowerPlex® ESI 17 Pro System Solution for X.1 and X.3 Single Nucleotide Polymorphisms
Given that the SE33 primer pair in the PowerPlex® ESX 17 System was not susceptible to these mutations, we based the new PowerPlex® ESI 17 Pro primer pair on these existing primers. Because the PowerPlex® ESX 17 System SE33 primers demonstrate a high level of concordance with the Polymeropoulos primers (6) , the chance for any significant new discordance is minimized. PowerPlex® ESI 17 Pro primers were designed to yield amplicons of similar size as those currently generated with the PowerPlex® ESI 17 System, thus allowing the use of current panel and bin sets for data analysis.
Primer pairs for the remaining 15 autosomal loci and amelogenin were kept the same as in the current PowerPlex® ESI 17 System, as was the 5X Master Mix, thereby minimizing any potential for discordance at any other loci. The 23.2 allele in GT37190 that had previously been called off-ladder with PowerPlex® ESI 17 ran on-ladder and was correctly called as a 23.2 allele with the PowerPlex® ESI 17 Pro System (Figure 3). Five DNA samples that demonstrated this migration shift with the original PowerPlex® ESI 17 System (including one with a C to T mutation 60 bases downstream of the repeat) were tested with the PowerPlex® ESI 17 Pro System. All samples typed correctly without the migration shift, demonstrating the ability of this new SE33 primer pair to overcome these X.1 and X.3 SNPs.
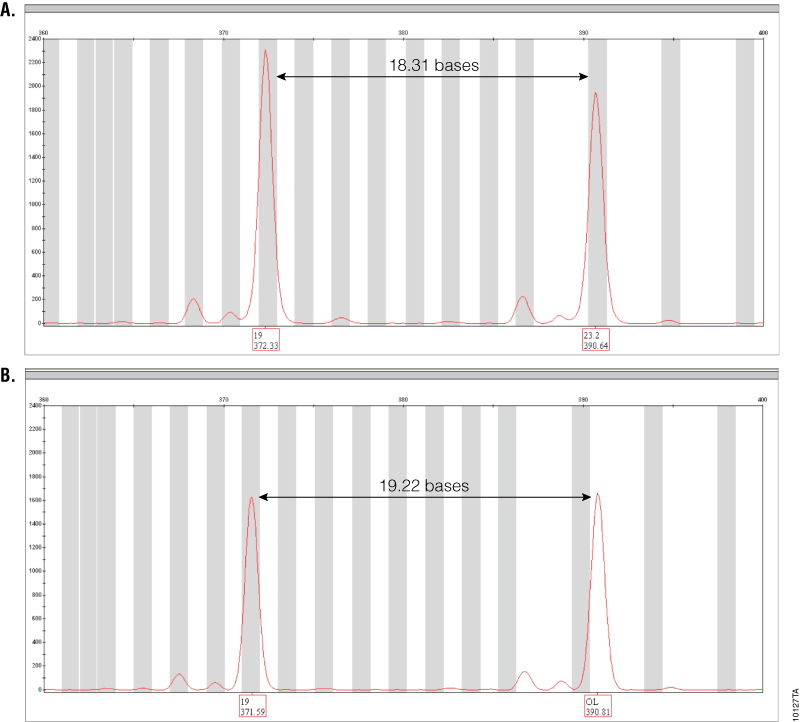 Figure 3. Amplification of GT37190 DNA with PowerPlex® ESI 17 Pro and ESI 17 Systems.
Figure 3. Amplification of GT37190 DNA with PowerPlex® ESI 17 Pro and ESI 17 Systems. The SE33 locus range is shown for both PowerPlex® ESI 17 Pro (Panel A) and ESI 17 (Panel B) amplifications of DNA sample GT37190. Peak labels show allele calls (top) and sizes in bases (bottom). The off-ladder peak seen with PowerPlex® ESI 17 is correctly called as 23.2 with the PowerPlex® ESI 17 Pro System.
The PowerPlex® ESI 17 Pro System Solution for Casework Artifacts
During the initial developmental validation work for the PowerPlex® ESI Systems, studies were conducted to confirm species specificity for these new multiplexes (5) . While these studies are useful in determining the likelihood of amplification from nonhuman DNA sources, they are unfortunately limited by the fact that it is impossible to model every possible organism that might be encountered in a casework setting. Recently, a few labs including LKA Mecklenburg-Vorpommern encountered casework samples that showed apparent nonhuman amplification peaks, specifically in the red dye channel, when using the PowerPlex® ESI 17 System. These background peaks were sometimes on-ladder and sometimes off-ladder but were eliminated when these samples were amplified with the PowerPlex® ESI 16 System, which lacks the SE33 primer pair. As similar reports of background peaks had not been reported with the PowerPlex® ESX 17 System, we expected the use of the new SE33 primer pair in the PowerPlex® ESI 17 Pro System, which is based on the PowerPlex® ESX SE33 primer pair, to eliminate these background peaks. A casework sample with a prominent artifact at 121.5 bases (as well as several other amplification artifacts below 50RFU in the red dye channel) in the D8S1179 locus when amplified with the PowerPlex® ESI 17 System, but not with the PowerPlex® ESI 16 System, was amplified side by side with the PowerPlex® ESI 17 and ESI 17 Pro Systems. While the peak is clearly present with PowerPlex® ESI 17, it is absent in the PowerPlex® ESI 17 Pro amplification (Figure 4).
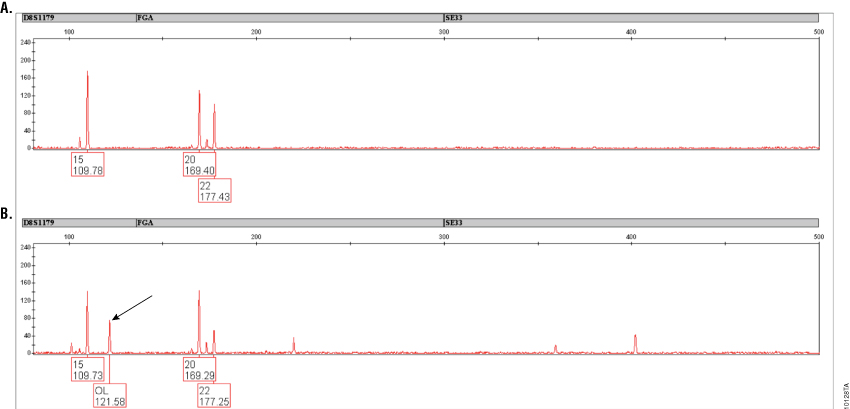 Figure 4. Amplification of a casework sample with PowerPlex® ESI 17 and ESI 17 Pro Systems.
Figure 4. Amplification of a casework sample with PowerPlex® ESI 17 and ESI 17 Pro Systems. Results are shown for a casework sample (taped glove) that was previously shown to exhibit an off-ladder peak at approximately 121.5 bases in D8S1179 with PowerPlex® ESI 17 (indicated by arrow in Panel B). Amplification with PowerPlex® ESI 17 Pro (Panel A) eliminates this peak. Peaks above 50RFU are labeled and show allele call (top) and size in bases (bottom). Peaks present below 50RFU are not labeled. Variation in peak heights is due to the low-level nature of the DNA sample.
Conclusion
The new PowerPlex® ESI 17 Pro System incorporates changes to the SE33 primer pair based on the PowerPlex® ESX SE33 primer sequences. This change allows robust amplification of DNA samples containing the X.1 or X.3 SNP and generated amplicons that are not affected by the migration shift caused by these mutations. As the 3′ ends of these new primers have the exact same sequence as primers found in the PowerPlex® ESX 17 System, we expected minimal, if any, effect on concordance at the SE33 locus, given the high level of concordance between this primer set and the Polymeropoulos primers (6) . Finally, the changes to this primer pair eliminated nonspecific amplification seen in some casework samples with the PowerPlex® ESI 17 System. Therefore, the new PowerPlex® ESI 17 Pro System should allow easier data interpretation for casework samples, with minimal potential for discordance at SE33.
Acknowledgments
* This work was funded in part by the National Institute of Justice (NIJ) through an interagency agreement 2008-DN-R-121 with the NIST Office of Law Enforcement Standards. Points of view in this document are those of the authors and do not necessarily represent the official position or policies of the U.S. Department of Justice. Certain commercial equipment, instruments and materials are identified in order to specify experimental procedures as completely as possible. In no case does such identification imply a recommendation or endorsement by the National Institute of Standards and Technology nor does it imply that any of the materials, instruments or equipment identified are necessarily the best available for the purpose.
Article References
- Sprecher, C.J. et al. (2009) The PowerPlex® ESX and ESI Systems: Meeting the new European standard. Profiles in DNA.
- Gill, P. et al. (2006) The evolution of DNA databases—Recommendations for new European STR loci. Forensic Sci. Int. 156, 242–4.
- Gill, P. et al. (2006) New multiplexes for Europe—Amendments and clarification of strategic development. Forensic Sci. Int. 163, 155–7.
- Tucker, V.C. et al. (2012) Developmental validation of the PowerPlex® ESX 16 and PowerPlex® ESX 17 Systems. Forensic Sci. Int. Genet. 6, 124–31.
- Tucker, V.C. et al. (2011) Developmental validation of the PowerPlex® ESI 16 and PowerPlex® ESI 17 Systems: STR multiplexes for the new European standard. Forensic Sci. Int. Genet. 5, 436–48.
- Hill, C.R. et al. (2011) Concordance and population studies along with stutter and peak height ratio analysis for the PowerPlex® ESX 17 and ESI 17 Systems. Forensic Sci. Int. Genet. 5, 269–75.
- Oldroyd, N. et al. (2011) Development of the AmpFlSTR® NGM SElect™ Kit: New sequence discoveries and implications for genotype discordance. Forensic News [Internet] [cited: 2011, December, 2].
- Polymeropoulos, M.H. et al. (1992) Tetranucleotide repeat polymorphism at the human beta-actin related pseudogene H-beta-Ac-psi-2 (ACTBP2). Nucleic Acid Res. 20, 1432.
- Wang, D.Y. et al. (2011) Identification and secondary structure analysis of a region affecting electrophoretic mobility of the STR locus SE33. Forensic Sci. Int. Genet. 6, 310–6.
- McLaren, R.S. et al. (2008) Post-injection hybridization of complementary DNA strands on capillary electrophoresis platforms: A novel solution for dsDNA artifacts. Forensic Sci. Int. Genet. 2, 257–73.
How to Cite This Article
Scientific Style and Format, 7th edition, 2006
McLaren, R.S. et al. Improved Primer Pair for the SE33 Locus in the PowerPlex® ESI 17 Pro System. [Internet] 2012. [cited: year, month, date]. Available from: https://www.promega.com/es-es/resources/profiles-in-dna/2012/improved-primer-pair-for-the-se33-locus-in-the-powerplex-esi-17-pro-system/
American Medical Association, Manual of Style, 10th edition, 2007
McLaren, R.S. et al. Improved Primer Pair for the SE33 Locus in the PowerPlex® ESI 17 Pro System. Promega Corporation Web site. https://www.promega.com/es-es/resources/profiles-in-dna/2012/improved-primer-pair-for-the-se33-locus-in-the-powerplex-esi-17-pro-system/ Updated 2012. Accessed Month Day, Year.
Contribution of an article to Profiles in DNA does not constitute an endorsement of Promega products.
PowerPlex is a registered trademark of Promega Corporation.
AmpFlSTR is a registered trademark of Life Technologies. NGM SElect and SEfiler Plus are trademarks of Life Technologies.
Products may be covered by pending or issued patents or may have certain limitations. More information.
All prices and specifications are subject to change without prior notice.
Product claims are subject to change. Please contact Promega Technical Services or access the Promega online catalog for the most up-to-date information on Promega products.