A Head-to-Head Comparison of RNasin® and RNaseOUT™ Ribonuclease Inhibitors
Promega Corporation
Publication Date: 2011
Abstract
In this article, we compared the performance of Recombinant RNasin® Ribonuclease Inhibitor to Invitrogen Corporation’s RNaseOUT™ Recombinant Ribonuclease Inhibitor. Both ribonuclease inhibitors were tested for their ability to protect RNA from degradation during three common applications: in vitro transcription, in vitro translation and qRT-PCR. In all three assays, Recombinant RNasin® Ribonuclease Inhibitor protected RNA from RNase A degradation better than RNaseOUT™ Recombinant Ribonuclease Inhibitor.
Introduction
Many applications in biochemistry and molecular biology require intact RNA, such as in vitro translation, in vitro transcription and qRT-PCR. Ribonucleases (RNases) are ubiquitous and can be introduced into experiments in many ways: from bare hands, pipettors and even the air. Many laboratory buffers, believed to be RNase-free, are actually contaminated with RNases (1) . For many reactions, RNase contamination can go unnoticed; however for RNA-sensitive applications, even a small amount of RNase can be detrimental to the experimental outcome. There are numerous commercially available RNase inhibitors designed to protect sensitive, RNA-based experiments. Placental ribonuclease inhibitor is a 50kDa protein that inhibits RNase by binding it in a 1:1 ratio with an association constant greater than 1016 M–1 (2) . It is expressed as a single-chain polypeptide consisting of 460 amino acid residues and contains leucine-rich repeats, a motif commonly associated with protein:protein interactions. The Promega Recombinant RNasin® Ribonuclease Inhibitor (Cat.# N2511) is a noncompetitive inhibitor of RNases A, B and C, human placental RNase and angiogenin. Although Recombinant RNasin® Ribonuclease Inhibitor affects a broad spectrum of RNases, it does not inhibit other nucleases, reverse transcriptases or polymerases (3) (3) . Additionally, Recombinant RNasin® Ribonuclease Inhibitor must pass quality control tests for contaminating RNase activity including latent RNases, DNase activity and endonuclease activity. Previously, RNasin® Ribonuclease Inhibitor was compared to Ambion’s SUPERase•IN™ and showed superior performance in total yeast RNA, in vitro transcription/translation and nondenaturing gel assays (3) . This article compares the performance of Promega Recombinant RNasin® Ribonuclease Inhibitor to Invitrogen’s RNaseOUT™ Recombinant Ribonuclease Inhibitor.
"For many reactions, RNase contamination can go unnoticed; however for RNA-sensitive applications, even a small amount of RNase can be detrimental to the experimental outcome."
Methods
RNase inhibitors
In all the experiments, we compared our Recombinant RNasin® Ribonuclease Inhibitor (Cat.# N2511) to the Invitrogen RNaseOUT™ Recombinant Ribonuclease Inhibitor. Both inhibitors are supplied at 40 units/µl with one unit defined as the amount of inhibitor required to inhibit the activity of 5ng of ribonuclease A (RNase A) by 50%. RNase A was purchased from Fermentas (Cat.# EN0531) and diluted to 10ng/µl.
In vitro Translation
Both TnT® T7 Quick Coupled Transcription/Translation System (Cat.# L1170) and an uncoupled Rabbit Reticulocyte Lysate System, Nuclease Treated (Cat.# L4960), were used to express firefly luciferase. Reactions for the TnT® T7 Quick Coupled System were assembled as instructed in the TnT® Quick Coupled Transcription/Translation Systems Technical Manual #TM045 using 2µl of Luciferase Control DNA as the template. Reactions for the Rabbit Reticulocyte Lysate System were assembled as instructed in the Rabbit Reticulocyte Lysate System Technical Manual #TM232 using 2µl of Luciferase Control RNA as the template. All reactions were performed in triplicate. For the reactions containing RNase A, 10ng were added. For reactions with an RNase inhibitor, 8 units were added. Reactions were incubated at 30°C for 90 minutes. After incubation, 2.5µl of each reaction was added to a 96-well plate along with 50µl of Luciferase Assay Reagent (Cat.# E1483). Luminescence was measured using the GloMax®-Multi+ Detection System.
In vitro Transcription
Riboprobe® System—T7 (Cat.# P1440) was used for all in vitro transcription reactions. Reactions were assembled as instructed in the Riboprobe® in vitro Transcription Systems Technical Manual #TM016 using the pGEM® Express Positive Control Template. All reactions were performed in duplicate. For the reactions containing RNase A, 10ng was added. For reactions with an RNase inhibitor, 50 units were added. Reactions were incubated at 37°C for 60 minutes. After incubation, 5µl of the reaction was mixed with 15µl of Formaldehyde Sample Buffer (Lonza Cat.# 50571) and incubated at 70°C for 5 minutes. Twenty microliters of each sample was loaded on a 1.2% agarose gel with 10µg/ml ethidium bromide and run at 100V for 30 minutes.
qRT-PCR
RT-qPCR reactions. Reactions were assembled using 1.2kb Kanamycin Positive Control RNA as template with the Kanamycin 25X primers. All reactions were performed in triplicate. For the reactions containing RNase A, 10ng was added. For reactions with an RNase inhibitor, 50 units were added. Reactions were amplified and detected using the CFX96™ Real-Time PCR Detection System.
Results
In vitro Translation
We tested both Recombinant RNasin® Ribonuclease Inhibitor and RNaseOUT™ Recombinant Ribonuclease Inhibitor for the ability to protect RNA template from RNase A degradation during in vitro translation. Luciferase was translated using both the Rabbit Reticulocyte Lysate System (Cat.# L4960; Figure 1, Panel A) and the TnT® T7 Quick Coupled Transcription/Translation System (Cat.# L1170; Figure 1, Panel B) in the presence of inhibitor alone and inhibitor plus RNase A. In both systems, RNase A alone reduced luminescence to less than 2% of the mRNA only control. Adding Recombinant RNasin® Ribonuclease Inhibitor did not decrease light production; however, RNaseOUT™ Recombinant Ribonuclease Inhibitor alone reduced light output to 83% for the Rabbit Reticulocyte Lysate System and 91% for the TnT® T7 Quick Coupled Transcription/Translation System. Additionally, RNaseOUT™ Recombinant Ribonuclease Inhibitor conferred only partial protection of RNA when RNase A was added to the reaction, measuring only 44% of the positive control signal in both systems. Recombinant RNasin® Ribonuclease Inhibitor performed much better with recovery to ~85% of the positive control signal in both cell-free expression systems.
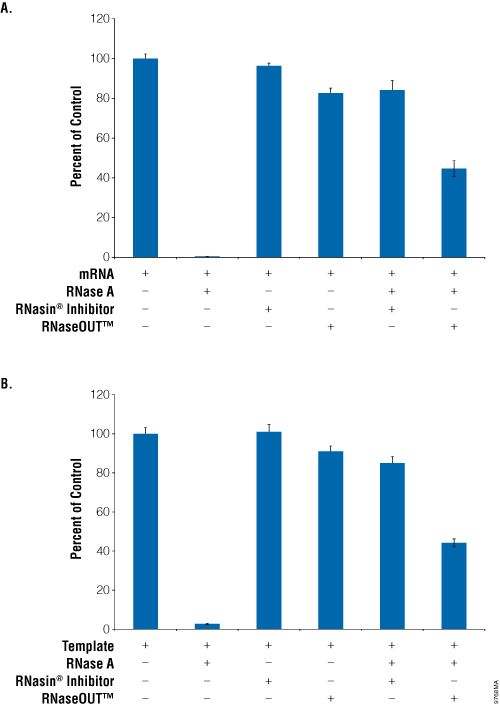 Figure 1. Comparison of RNasin® Ribonuclease Inhibitor and RNaseOUT™ inhibition of RNase A during in vitro translation.
Figure 1. Comparison of RNasin® Ribonuclease Inhibitor and RNaseOUT™ inhibition of RNase A during in vitro translation.
Panel A. Luciferase mRNA was translated in Rabbit Reticulocyte Lysate, Nuclease Treated, per Technical Manual #TM232. Panel B. Luciferase DNA was transcribed and translated in the TnT® T7 Coupled Transcription/Translation System as instructed in Technical Manual #TM045. Luciferase activity was assessed using the Luciferase Assay System and luminescence detected using a GloMax®-Multi+ Detection System with Instinct™ Software. Percent of Control was calculated as a percent light produced compared to the positive control (template only). Error bars represent three separate reactions.
In vitro Transcription
To compare the ability of Recombinant RNasin® Ribonuclease Inhibitor and RNaseOUT™ Recombinant Ribonuclease Inhibitor to protect RNA product during in vitro transcription, we used the Riboprobe® System—T7 (Cat.# P1440) with the pGEM® Express Positive Control Template. The template was transcribed in the presence of inhibitor alone and inhibitor plus RNase A, and reaction products were separated on a 1.2% agarose gel. Bands were quantitated using Image J 1.44 software from the National Institutes of Health. The template produced two products, one at 2,346 bases and one at 1,056 bases (Figure 2). The DNA template was observed in all lanes between 9.0kb and 5.0kb as determined by a polymerase only control. RNA was absent from the template only control and in the RNase A lanes. Similar RNA intensities were observed in the control lanes (template + polymerase), Recombinant RNasin® Ribonuclease Inhibitor only, and RNaseOUT™ Recombinant Ribonuclease Inhibitor only lanes. When RNase A and inhibitor were added to the reaction, both band intensities were reduced; however, the Recombinant RNasin® Inhibitor reaction produced bands corresponding to the 2,346bp product that were measured as 38.5% of the control, while RNaseOUT™ Recombinant Ribonuclease Inhibitor reactions produced bands only 9.8% of the control. More full-length products were seen in the RNasin® Ribonuclease Inhibitor/RNase A lanes than in the RNaseOUT™ inhibitor/RNase A lanes, indicating that Recombinant RNasin® Ribonuclease Inhibitor inhibited RNase A better than RNaseOUT™ Recombinant Ribonuclease Inhibitor.
 Figure 2. Comparison of RNasin® Ribonuclease Inhibitor and RNaseOUT™ inhibition of RNase A during in vitro transcription.
Figure 2. Comparison of RNasin® Ribonuclease Inhibitor and RNaseOUT™ inhibition of RNase A during in vitro transcription. pGEM® Express Control Template was transcribed using the RiboProbe® System—T7 following the instructions in Technical Manual #TM016. All reactions were performed in duplicate. Expected sizes in bases are displayed on the right of the gel image, and marker sizes in kb are listed on the left. The graph was generated by quantitating the bands on the gel and calculating a percent RNA compared to the positive control (polymerase + template). Error bars represent the two separate reactions. The black bars are quantitation of the top bands (2,346 bases) and the grey bars are quantitation of the bottom bands (1,065 bases).
qRT-PCR
Real-time amplification is a powerful tool in both detection and quantification of one or more specific sequences in a DNA sample. The Plexor® One-Step qRT-PCR System is a novel real-time PCR system for the quantitation of specific sequences within an RNA sample. This one-tube, two-enzyme system uses ImProm-II™ Reverse Transcriptase and the Plexor® qPCR technology for quantitation of any type of RNA sample. Having intact RNA is essential for detection and quantification of any sample used for qRT-PCR. To compare the inhibitors' abilities to protect RNA product during qRT-PCR, we used the Plexor® One-Step qRT-PCR System (Cat.# A4021) using the 1.2kb Kanamycin Positive Control RNA for the template. Samples were amplified with RNase A only, RNase inhibitor only and a combination of RNase A and RNase inhibitor. The data were analyzed using Plexor® software and reported in percent mRNA (Figure 3). Percent-mRNA values were derived by inserting the cycle threshold (Ct) values into the equation 2ΔCt × 100, where delta Ct is the difference between the positive control (mRNA only) and the experimental Ct values. With only RNase A added, no mRNA was detected. This indicates that 10ng of RNase A can completely degrade all the RNA in the sample. When adding either Recombinant RNasin® Ribonuclease Inhibitor only or RNaseOUT™ Recombinant Ribonuclease Inhibitor only, the mRNA values were 113% and 103%, respectively, indicating these inhibitors had no negative effects on the reverse transcription and PCR steps. When both RNase A and RNase inhibitor were added, the Recombinant RNasin® Ribonuclease Inhibitor reactions showed mRNA values of 115%, whereas RNaseOUT™ Recombinant Ribonuclease Inhibitor gave a value of 34%. Recombinant RNasin® Ribonuclease Inhibitor inhibited RNase A better than RNaseOUT™, ensuring the RNA was reverse transcribed and amplified.
 Figure 3. Comparison of RNasin® Ribonuclease Inhibitor and RNaseOUT™ inhibition of RNase A during qRT-PCR.
Figure 3. Comparison of RNasin® Ribonuclease Inhibitor and RNaseOUT™ inhibition of RNase A during qRT-PCR.
Kanamycin Control RNA was reverse transcribed and amplified using the Plexor® One-Step qRT-PCR System according to Technical Manual #TM263. All reactions were performed in triplicate. Graph was generated from the following formula: 2ΔCt × 100. Error bars represent three separate reactions.
Conclusions
RNases are ubiquitous and can have profound effects on data in downstream applications. RNase inhibitors allow researchers to safeguard against RNase contamination in their experiments. Applications such as in vitro transcription, in vitro translation and qRT-PCR require intact RNA template for successful results. We compared two RNase inhibitors, Recombinant RNasin® Ribonuclease Inhibitor and RNaseOUT™ Recombinant Ribonuclease Inhibitor, in three different types of experiments and found Recombinant RNasin® Ribonuclease Inhibitor outperformed RNaseOUT™ Recombinant Ribonuclease Inhibitor in protecting RNA in all three experiments.
Article References
- Hendricksen, A., Hook, B. and Schagat, T. (2010) RNase contamination happens; Recombinant RNasin® Inhibitor can safeguard your samples. Promega PubHub.
- Dickson, K.A., Haigis, M.C. and Raines, R.T. (2005) Ribonuclease inhibitor: Structure and function. Prog. Nucleic Acid Res. Mol. Biol. 80, 349–74.
How to Cite This Article
Scientific Style and Format, 7th edition, 2006
Hook B, Hendricksen A and Schagat T. A Head-to-Head Comparison of RNasin® and RNaseOUT™ Ribonuclease Inhibitors. [Internet] 2011. [cited: year, month, date]. Available from: https://www.promega.com/resources/pubhub/head-to-head-comparison-of-rnasin-to-rnaseout/
American Medical Association, Manual of Style, 10th edition, 2007
Hook B, Hendricksen A and Schagat T. A Head-to-Head Comparison of RNasin® and RNaseOUT™ Ribonuclease Inhibitors. Promega Corporation Web site. https://www.promega.com/resources/pubhub/head-to-head-comparison-of-rnasin-to-rnaseout/ Updated 2011. Accessed Month Day, Year.
GloMax, pGEM, Plexor, Riboprobe, RNasin and TnT are registered trademarks of Promega Corporation. Instinct and ImProm-II are trademarks of Promega Corporation.
CFX96 is a trademark of Bio-Rad Laboratories, Inc. RNaseOUT is a trademark of Invitrogen Corporation. SUPERase•IN is a trademark of Ambion, Inc.
Products may be covered by pending or issued patents or may have certain limitations. Please visit our web site for more information.