Viral Research
We offer collaborative support and a broad portfolio of reagents that are used in research labs studying coronaviruses and other emerging viral diseases. We support scientists working to develop vaccines and to answer questions about viral pathology and treatment including:
- How does the virus enter human cells?
- How does the virus make people sick?
- What treatments can be used to alleviate symptoms?
- How can immunity to the virus be gained?
SARS-CoV-2 Drug Discovery and Vaccine Development
Assays and technologies for SARS-CoV-2 include solutions for monitoring SARS-CoV-2 viral entry, detecting neutralizing antibodies and their ADCC or ADCP activity, and measuring viral protease activity.
Virus:Host Cell Interactions
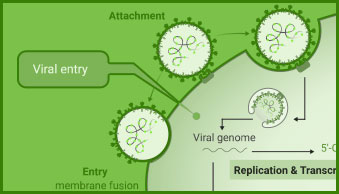
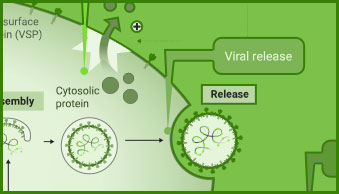
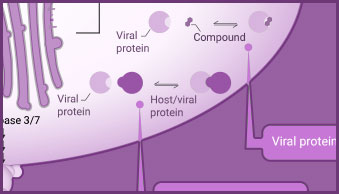
Interaction between viral and host cell proteins is fundamental for the viral replication cycle to proceed. These include interaction with host cell surface proteins that function as viral entry receptors, as well as the various interactions taking place intracellularly upon viral entry. These virus:virus and virus:host-cell interactions are important targets for therapeutic intervention. Vaccine effectiveness can also be determined by examining the ability of immune serum to prevent the interactions necessary for cellular entry of the virus.
NanoBiT® and NanoBRET® Technologies, both based on NanoLuc® Luciferase, enable live-cell analysis of protein interactions. Together with firefly luciferase, NanoLuc Luciferase is utilized for the generation of reporter viruses, pseudotyped virus particles and pseudotyped virus-like particles to study viral entry and release.
The versatile HiBiT Protein Tagging System allows for monitoring the dynamics of viral infection in real time. The small size of the HiBiT reporter tag (11 aa; 1.3 kDa) makes it particularly useful for viral studies because it can be easily integrated into small viral genomes.
Products for Monitoring Virus:Host Cell Interactions
Protein Interaction Assays
Products for the analysis of viral:host protein interactions include NanoBiT® complementation assay, NanoBRET® BRET-based assays and pull-down assays.
Protein Quantitation Assays
Tag proteins with the 11aa HiBiT tag and detect with reagents containing the high-affinity LgBiT subunit to form a bright luminescent signal.
Luciferase Reporter Assays
Multiple options for detecting NanoLuc®, firefly, or Renilla luciferase to create reporter assays optimized for specific experimental goals. Both lytic or live-cell options as well as single- or dual-reporter detection.
Virus:Host Interaction Resources
Host Immune Response
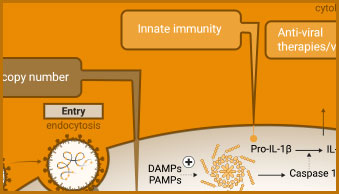
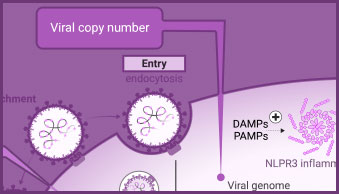
The innate immune response includes activation of inflammasomes and cytokine release. Caspase-1 activity, the release of cytokines (e.g., IL-1-β) or damage-associated molecular patterns (DAMPs) can all be used to evaluate aspects of the innate immune response in infected cells.
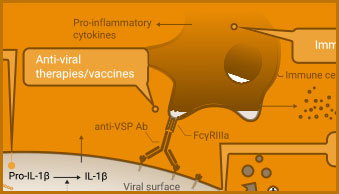
The adaptive immune response includes production of neutralizing antibodies and antibodies with ADCC activity, in addition to T cell activation. Promega’s portfolio of bioluminescent reporter assays supports the development of vaccines to induce antibody response. Cytokine detection assays can be used to monitor T cell activation.
Products for Monitoring Host Immune Response
Caspase-1 Activity
Conversion of procaspase-1 zymogen into active caspase-1 is a key marker of inflammasome formation.
Inflammation Assays
We offer simple add-mix-measure assays for the detection of a variety of cytokines in cell culture samples.
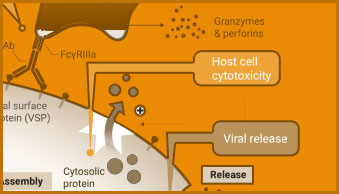
ADCC Activity Bioassay
The ADCC Reporter Bioassay can be used to measure the potency and stability of antibodies that bind and activate Fcγ receptors.
Monitor Host Cell Response
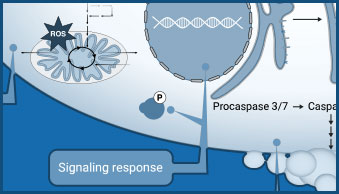
Viral infection can lead to a multitude of different host cell responses. These range from changes in gene expression, signaling, and metabolism to cytopathic effects that alter cell health. These virus-induced changes can be understood using assays that monitor nutrient uptake or consumption and changes in co-factor levels.
Screening compounds for antiviral activity includes testing for viral-induced cytopathic effects (CPE) in host cells. Cell-based viability and cytotoxicity assays such as the CellTiter-Glo® Cell Viability Assay are amenable to high-throughput analysis, reducing the time needed to analyze the effect of potential treatments. Viability and cytotoxicity assays can also be used to support studies investigating the mechanism of action of viruses.
Using Cell Viability Assays in Screens for SARS-CoV-2 Drug Candidates
How the coronavirus enters cells and how to block it
Read about viral host interactions, and how cell viability assays support these studies.
Dr. Colleen Jonsson, Director of the Regional Biocontainment Laboratory and Director of the Institute for the Study of Host-Pathogen Interactions at the University of Tennessee Health Science Center, has been studying highly pathogenic human viruses for more than three decades. She has led several cross-institutional projects using high-throughput screens to discover small molecules that could be used as antiviral drugs. And now, she’s using that experience to find an antiviral therapeutic against SARS-CoV-2. For her high-throughput screens, Dr. Jonsson used the CellTiter-Glo® Cell Viability Assay to determine whether the small molecules inhibit virus-induced cell death. Read more here.
“The CellTiter-Glo® Assay provides a very robust, easy-to-use reagent for high-throughput screening of small molecules for antiviral activity.”
Products for Monitoring Host Cell Response
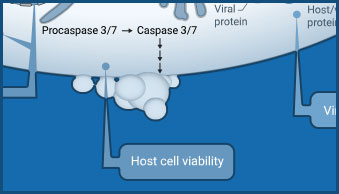
Host Cell Viability
CellTiter-Glo® products provide a simple "add-mix-measure" protocol for determining cell viability.
Viral Toxicity
A simple, quantifiable method for determining viral-induced cytopathic effects (CPE) in host cells caused by lytic virions:
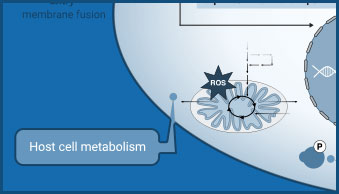
Host Cell Metabolism
Lactate assays can be used to detect changes in glycolysis as a result of viral infection.
Detect and Monitor Viral Genome Copy Number
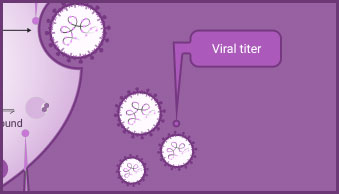
PCR-based methods, such as endpoint and real-time PCR and RT-PCR, quantify changes in viral genome copy number to signify viral replication/proliferation. They are fundamental tools used in the development of viral detection tests, for analysis of viral genomes, and for measuring changes in viral genome copy number as a surrogate assay for viral replication.
GoTaq® PCR and qPCR systems and GoScript™ Reverse Transcriptase offer robust and reproducible amplification and reverse transcription for detecting and amplifying sequences for numerous viral targets.
Products for Monitoring Viral Replication
Ready-to-Use Master Mixes
Ready-to-use mix of optimized components for robust qPCR and RT-qPCR using hydrolysis probes.
PCR Optimization
Preformulated PCR master mix components to determine optimal PCR amplification conditions.
Reverse Transcription
Reliable cDNA synthesis for sensitive detection of expression. Available as a standalone enzyme, a complete kit or as a master mix with oligo(dT) or random primers.
RNA Production for Vaccine Research and Development
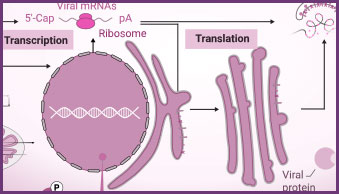
Transcribed RNA is required for vaccine production, viral standards, and basic viral research. For in vivo and in vitro studies, RiboMAX® RNA Production System generates a large quantity of high-quality RNA or mRNA from a DNA template without the need for mammalian cells or cell components. These in-vitro transcribed viral RNA or mRNA transcripts, typically encoding a disease-specific antigen such as the spike protein of a coronavirus, may be used as inoculation material for viral infection studies. If the transcribed mRNA is to be used as a therapeutic, the mRNA encoding a desirable protein can be packaged as necessary for delivery to the tissue of interest.

RNA Production Products and Resources
RNA Production Product
RiboMAX™ in vitro transcription system produces 2–5mg/ml RNA in a 1ml reaction.