Screening for compounds that inhibit growth and sporulation of fungi and growth of algae using a luminescent assay
Janssen PMP, Turnhoutseweg 30, B-2340 Beerse, Belgium
Publication Date: 2011
Introduction
Biological assays involving germination or growth are often used to determine viability of fungal spores (1). However many fungal spores, such as those from soil-inhabiting basidiomycetes, are difficult to germinate in the laboratory, and some fungal species require months to germinate (1,2). Other fungal viability assays involve the use of colorimetric assays such as MTT/XTT assays (3,4) or fluorescent stains (5,6). Many fluorescent stains are subject to fluorescence interference from test compounds and may not be ideal for screening situations (7). These factors make activities like screening for sporicides almost impossible. A rapid viability test that works well with fungal spores would be useful for screening for chemicals that inhibit sporulation.
Determining algal viability can be extremely difficult as well, often requiring labor-intensive and expensive spectroscopy methods or methods such as measuring photosynthetic oxygen that are not well-suited to high-throughput screening (8–11). Fluorescent stains have been widely applied in studies of algal viability, but these studies are subject to interference from green autofluorescence that occurs in many species (9). As production of certain crops that require aquaculture (e.g., rice) becomes more prevalent, understanding the effect of herbicides used in those agricultural practices on the normal algal flora becomes important (10). Also, finding effective agents for controlling growth of harmful algal blooms requires being able to rapidly assess algal viability in a screening setting within the laboratory. Some studies have had success with MTS-based assays (12); however, a truly homogeneous assay that can be performed on organisms grown in culture in conjunction with screening assays would be a powerful tool.
In this report we demonstrate the use of the luminescent BacTiter-Glo™ Microbial Cell Viability Assay for screening of fungal sporicidal compounds and show that it can be used to assess viability of yeast and algae.
Assay Background
The BacTiter-Glo™ Microbial Cell Viability Assay provides a method for determining the number of viable bacterial cells in culture by measuring the amount of ATP (13). The “add-mix-measure” format of the assay allows users to simply add a reagent, mix and then measure the luminescent output of the assay. The reagent supports cell lysis and generation of a luminescent signal that is proportional to the amount of ATP present, which is directly proportional to the number of cells in culture (13).
The assay uses a proprietary reagent formulation containing a highly stabilized luciferase, Ultra-Glo™ Recombinant Luciferase and detergent, to extract ATP from bacterial cells and support a stable “glow-type” luminescent signal. Because the reagent is physically robust and the luminescent output is sensitive and stable the BacTiter-Glo™ Assay is well suited for screening applications that may require the processing of many multiwell plates.
To perform the assay, lyophilized BacTiter-Glo™ Substrate is reconstituted using the BacTiter-Glo™ Buffer and equilibrated to room temperature for 15 minutes. An equal volume of reagent is added to the microbial cell culture, mixed and incubated for 5 minutes. The emitted luminescence is detected using a luminometer.
The BacTiter-Glo™ Assay has been used successfully with Gram+ and Gram– bacteria and two species of yeast (13) and to screen a panel from the Library of Pharmacological Active Compounds (LOPAC) from Sigma for antimicrobial activity against Staphylococcus aureus (13). The assay has been used to study bacterial attachment in biofilm formation (14), to screen for inhibitors of Pseudomonas biofilm formation (15), and for the rapid counting of mycobacterium complex (MAC) organisms (16).
In this report, we demonstrate effectiveness of the BacTiter-Glo™ Assay in detecting microbial number of yeast, algae and fungi, and we use it in a test screen for sporicidal compounds against Mucor piriformis, a plant pathogen that causes rot of certain fruits such as apple and pear after harvest.
Results
Determining Growth of R. rubra using the BacTiter-Glo™ Assay
First we sought to demonstrate the utility of the BacTiter-Glo™ Assay to measure growth of the yeast Rhodotorula rubra strain IMI35841. Yeast cultures were grown overnight and serial dilutions were prepared. The BacTiter-Glo™ Reagent was prepared and assay performed as described in BacTiter-Glo™ Microbial Cell Viability Assay Technical Bulletin TB337. Figure 1 shows that the luminescent signal produced by the BacTiter-Glo™ Assay decreases upon dilution of R. rubra cells.
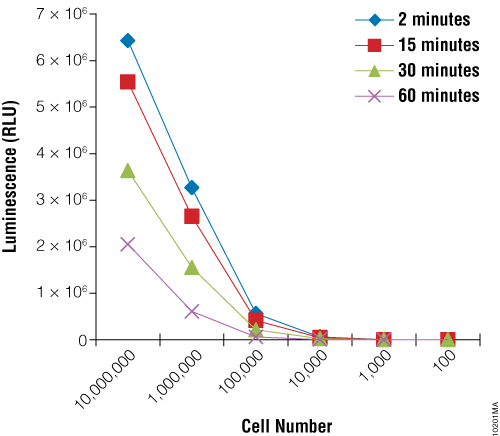
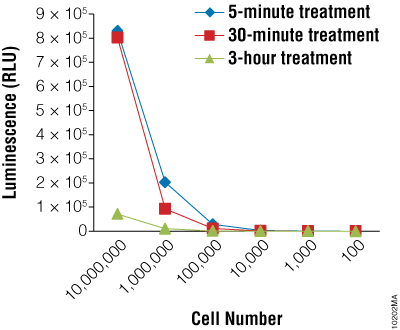
Next we wanted to determine if the BacTiter-Glo™ Assay could detect toxicity resulting from fungicide treatment of R. rubra cells. Cells were grown overnight as described for Figure 2 and a series of dilutions were made. Diluted cells were treated with the fungicide for 5, 30 or 180 minutes. Data indicate that the BacTiter-Glo™ Assay is suitable for detecting growth inhibition resulting from fungicidal compounds (Figure 2).
Using the BacTiter-Glo™ Assay to Detect Viability of Fungal Spores and Screen for Inhibitors of Sporulation
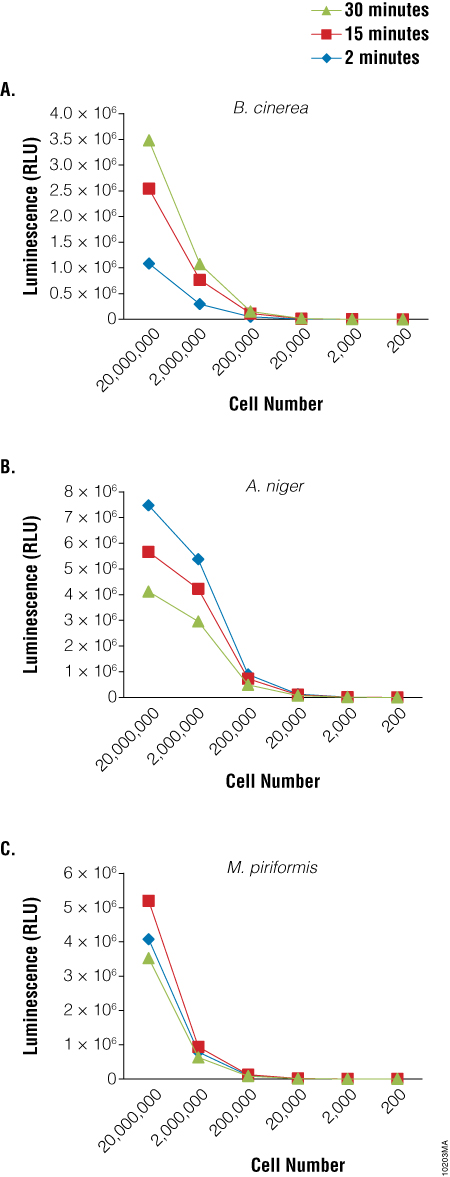
Because germination and growth assays are not particularly amenable to HTS screening activities, a rapid viability assay that could be used to screen for agents that inhibit sporulation would be particularly useful. We used the BacTiter-Glo™ Assay to detect the viability of spores of three fungal species: Botrytis cinerea, Aspergillus niger and Mucor piriformis. A clear relationship was observed between the luminescent output of the assay and the dilution of the spores (Figure 3). Similar results were obtained using spores of from several Penicillium species as well (data not shown). Experiments to determine viability of cells from fungal mycelia using the BacTiter-Glo™ Assay are ongoing.
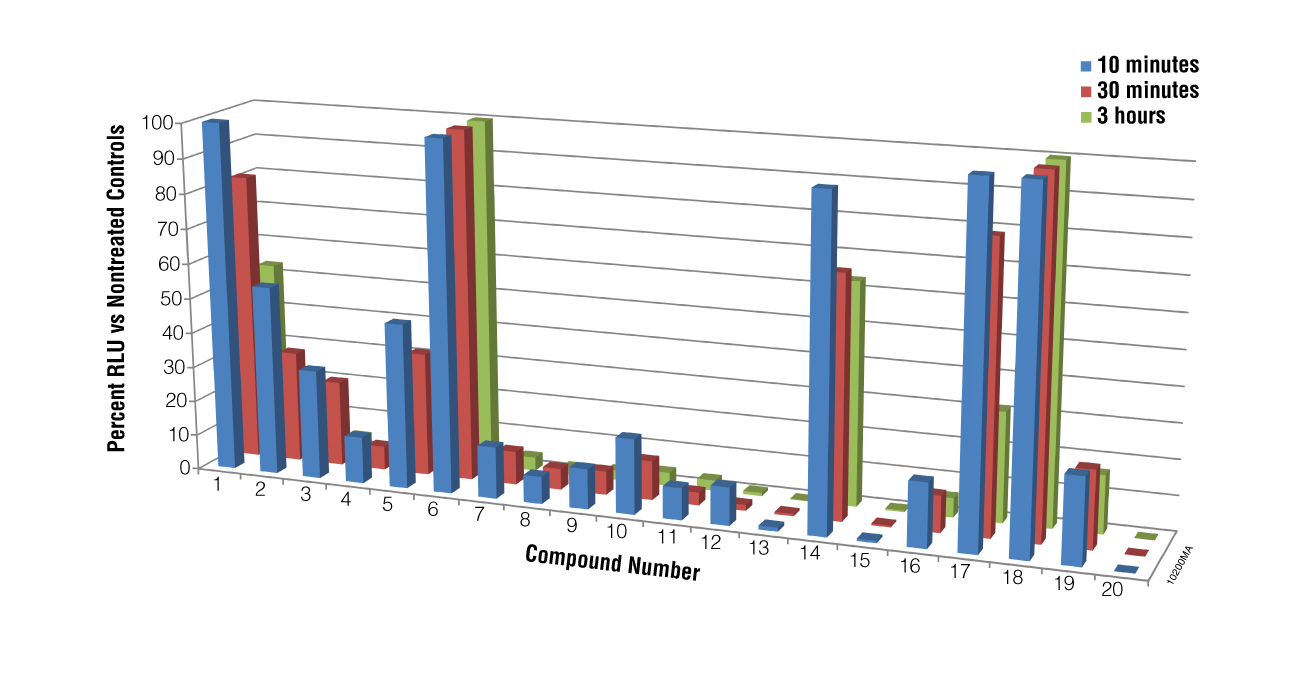
We next performed a screen of twenty compounds using M. piriformis spores as the test cells to determine if the BacTiter-Glo™ Assay could be used for screening potential compounds for sporicidal activity and to determine if the assay could distinguish a weak sporicidal agent from a strong one. Spores were collected and treated with compounds for 5, 30 or 180 minutes. While luciferase inhibition by the compounds cannot be excluded in the primary screen results for compounds 13, 15 and 20, when spores treated with these compounds were checked microscopically for growth at 24 hours, no germination was observed, indicating that the lack of luminescence was a result of compound action on the spores, not luciferase inhibition. The data presented in Figure 4 show that the BacTiter-Glo™ Assay was able to distinguish weak from strong sporulation inhibitors.
Using the BacTiter-Glo™ Assay with Algal Cell Culture
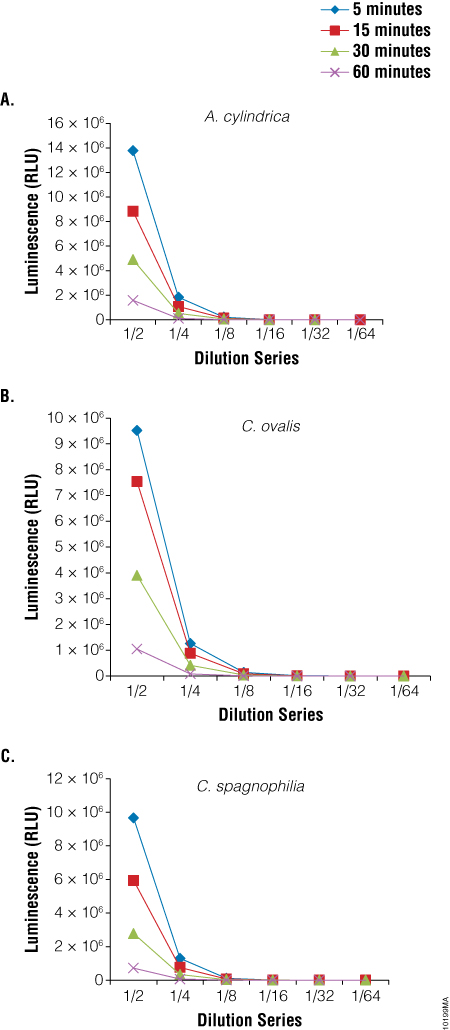
We also used the BacTiter-Glo™ Assay to assess viability in three species of algae: Anabaena cylindrica, a filamentous algae, Chlorella ovalis and Chlamydomonas spagnophilia, both unicellular algae. Algae were cultured in Blue-Green Medium (BG11 Broth). Serial dilutions of the cultures were made before performing the BacTiter-Glo™ Assay. Luminescence output from the assay decreased with the decrease in cell number expected with dilution (Figure 5).
Summary
Here we have demonstrated the utility of the BacTiter-Glo™ Microbial Cell Viability Assay for assessing metabolically active fungal and algal cells. We have demonstrated the ability of the assay to detect toxicity in one yeast strain from treatment with a fungicide. Furthermore, we have presented data that indicate the assay can be used to screen compounds for fungal sporicidal activity and that it can distinguish between weak and strong sporicidal compounds. This rapid assay for spore viability provides an essential tool for discovering new and effective fungal sporicidal compounds.
Featured Product
References
- Yu, S.Q., Trione, E.J. and Ching, T.M. (1984) Biochemical determination of the viability of fungal spores and hyphae Mycologia 76, 608–13.
- Fries, N. (1978) Basidiospore germination in some mycorrhiza-forming Hymenomycetes. Trans. Brit. Mycol. Soc. 70, 319–24.
- Levitz, S.M. and Diamond, R.D. (1985) A rapid colorimetric assay of fungal viability with the tetrazolium salt MTT. J. Infect. Dis. 152, 938–45.
- Tova, M. et al. (1995) A simplified new assay for assessment of fungal cell damage with the tetrazolium dye, (2,3)-bis-(2-methoxy-4-nitro-5-sulfophenyl)-(2H)-tetrazolium-5-carboxanilide (XTT). J. Infect. Dis. 172, 1153–6.
- Bowman, J.C. et al. (2002) The antifungal echinocandin caspofungin acetate kills growing cells of Aspergillus fumigatus in vitro. Antimicrob. Agents Chemother. 46, 3001–12.
- Lopes, M.A. et al. (2003) Fluorescent method for studying morphogenesis and viability of dermatophyte cells. Mycopathologica 156, 61–6.
- Thorne, N., Auld, D.S. and Inglese, J. (2010) Apparent activity in high-throughput screening: origins of compound-dependent assay interference. Curr. Opin. Chem. Biol. 14, 315–24.
- Gilboa, A. and Ben-Amotz, A. (1979) An improved method for rapid assaying of viability of cryopreserved unicellular algae. Plant Science Letters 14, 317–20.
- Tang, Z-Y. and Dobbs, F.C. (2007) Green autofluorescence in dinoflagellates, diatoms, and other microalgae and its implications for vital staining and morphological studies. Appl. Environ. Microbiol. 73, 2306–13.
- Nagi, T. et al. (2011) Effects of four rice paddy herbicides on algal cell viability and the relationship with population recovery. Environ. Toxicol. Chem. 30, 1898–905.
- Nasse, M. et al. (2009) Demountable liquid/flow cell in vivo infrared microspectroscopy of biological specimens. Appl. Spectrosc. 63, 1181–6.
- Capasso, J.M. et al. (2003) A colorimetric assay for determination of cell viability in algal cultures. Biomol. Eng. 20, 133–8.
- Fan, F. et al. (2004) Determining microbial viability using a homogeneous luminescent assay. Cell Notes 10, 2–5.
- Sule, P. et al. (2008) Use of the BacTiter-Glo™ Microbial Cell Viability Assay to study bacterial attachment in biofilm formation. Promega Notes 99, 19–21.
- Junker, L.M. and Clardy, J. (2007) High-throughput screens for small-molecule inhibitors of Pseudomonas aeruginosa biofilm development. Antimicrob. Agents Chemother. 51, 3582–90.
- Yuroff, A. et al. (2008) Application of BacTiter-Glo™ Assay for enumeration and screening of antimicrobial compounds for Mycobacterium avium. Promega Notes 98, 8–10.