Overcoming Challenges of DNA Purification from FFPE Samples: The ReliaPrep™ FFPE gDNA Miniprep System
Doug Wieczorek, Trista Schagat and Eric Vincent
Promega Corporation
Publication Date: Oct. 2014 (tpub_154)
Abstract
Archives of formalin-fixed and paraffin-embedded tissue sections are a valuable tool to study diseases such as cancer, but DNA purification from these tissues can be difficult because the DNA is often highly cross-linked, degraded and fragmented. The ReliaPrep™ FFPE gDNA Miniprep System has many unique features that can overcome these challenges in a safe, quick and effective manner.
Introduction
The combination of formalin fixation and paraffin embedding (FFPE) is a commonly used method for archiving pathological tissue specimens. Archives of FFPE tissue sections can be many years old and are a valuable tool to study diseases such as cancer. The ability to extract nucleic acid from FFPE tissue sections enables researchers to correlate the disease state and tissue morphology of a patient with a particular genetic trait. However, DNA extraction from these tissues is associated with a number of challenges, mainly due to the formalin fixation, which results in cross-linking between proteins and DNA as well as between different DNA strands. The end result is purified DNA that is often significantly degraded and fragmented. The degree of fragmentation depends on tissue type, age of the sample and specific conditions used during fixation. These factors can lead to issues with downstream applications, many of which incorporate PCR. Thus, the protocol for DNA purification from FFPE tissues must efficiently extract highly fragmented DNA and reverse cross-linking caused by formalin fixation.
The ReliaPrep™ FFPE gDNA Miniprep System is designed specifically for DNA extraction from FFPE samples. The workflow is shown in Figure 1. Typically, deparaffinization of FFPE sections is done with xylene or other harsh organic solvents. The ReliaPrep™ System uses nontoxic mineral oil to safely and effectively deparaffinize FFPE sections in a shorter period of time than organic solvents. The ReliaPrep™ System features optimized lysis conditions to reverse modifications introduced by formalin fixation without the need for overnight digestion. The RNase treatment occurs in the same lysis buffer, streamlining the preprocessing steps. Nucleic acid binding and washing occur on specially designed columns to capture partially degraded and fragmented DNA inherent to FFPE samples. As most FFPE samples are unique and often have limited amounts of tissue, the purified DNA can be eluted in as little as 30µl, providing a concentrated DNA sample for use in downstream applications where concentration may be critical to success.
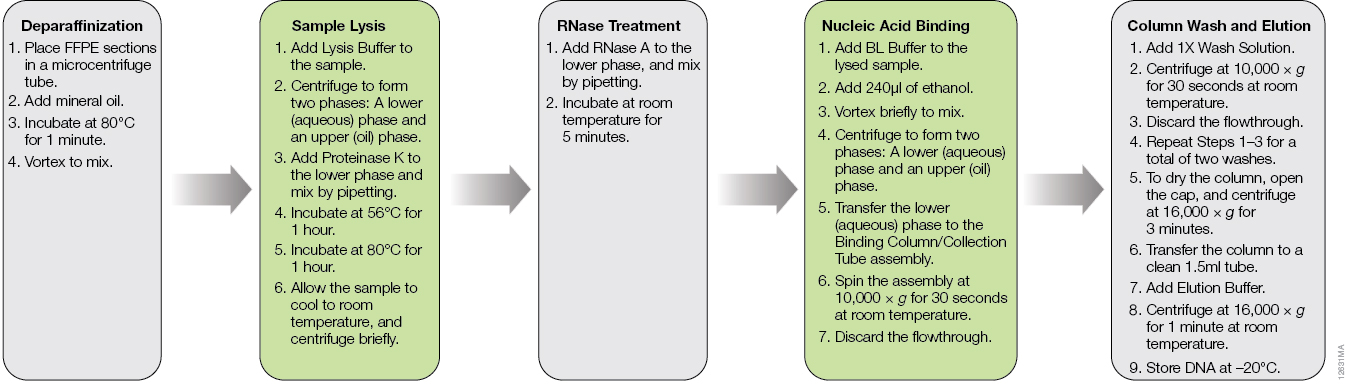
Figure 1. The ReliaPrep™ FFPE gDNA Miniprep System workflow.
Methods
Four FFPE tissues types were obtained from Biochain Institute, Inc. (Newark, CA): human lung tumor (Cat.# T2235152-D01), human breast tumor (Cat.# T2235086-D01), human colon tumor (Cat.# T2235090-D01) and human adult normal heart (Cat.# T2234122). DNA was purified from single 5µm FFPE tissue sections using three methods: 1) The ReliaPrep™ FFPE gDNA Miniprep System was used according to the standard protocol (1) , which includes a decross-linking step consisting of an 80°C incubation for 1 hour, 2) The ReliaPrep™ FFPE gDNA Miniprep System was used with a modified protocol, where the incubation time for decross-linking was increased to 4 hours and 3) The QIAamp® DNA FFPE Tissue Kit was used according to the standard protocol (2) , which includes a decross-linking step consisting of a 90°C incubation for 1 hour. For all methods, DNA was eluted in 30µl, with 30µl of eluate recovered.
DNA recovery was determined by qPCR analysis using primer sets that produced amplicons of varying sizes. As DNA purified from FFPE tissue is often degraded and highly fragmented, we chose amplicon sizes of less than 150bp in length for more accurate DNA quantitation. Amplification of larger amplicons will likely underestimate the available DNA. The GoTaq® qPCR Master Mix (dsDNA-binding dye-based qPCR, Cat.# A6001) was used with two primer sets specific to glyceraldehyde-3-phosphate dehydrogenase (GAPDH), resulting in 100bp and 300bp amplicons. Two microliters of DNA was added in a 25µl total reaction volume. The GoTaq® Probe qPCR Master Mix (fluorogenic probe-based qPCR, Cat.# A6101) was used with two primer sets specific to RNaseP and telomerase reverse transcriptase (TERT), resulting in 102bp and 164bp amplicons, respectively. Two microliters of DNA was added in a 20µl total reaction volume. For both master mixes, DNA was quantified using the Plexor® HY Male Genomic DNA Standard at amounts ranging from 100ng to 10pg. All samples were analyzed using the BioRad CFX 96 Real-Time PCR System.
Results and Discussion
Successful DNA purification and quantitation from FFPE samples is made difficult by the formalin-fixation process, which reduces the efficiency of subsequent amplifications and makes total yield estimates by qPCR unreliable. However, many downstream applications of DNA isolated from FFPE samples rely on amplification, so the amount of functional or amplifiable DNA versus the amount of total DNA can be an important distinction to make. We looked into whether increasing the decross-linking step from 1 hour to 4 hours would increase the amount of amplifiable DNA. A comparison of DNA recoveries from the four tissues using the three extraction methods, as determined using qPCR with each primer set, is shown in Figure 2.
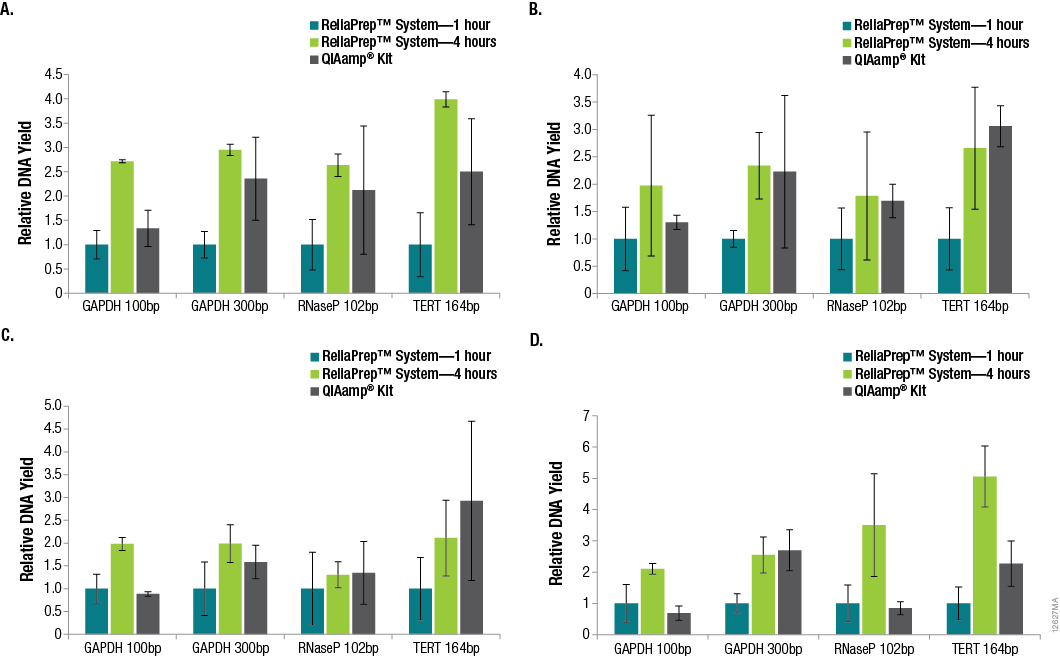
Figure 2. Relative DNA recoveries from FFPE tissues using three purification methods. Panel A. Lung tissue. Panel B. Breast tissue. Panel C. Colon tissue. Panel D. Heart tissue. Yields were determined using qPCR with primer sets that resulted in varying amplicon lengths. Yields are relative to those obtained using the standard ReliaPrep™ FFPE gDNA Miniprep System protocol (1-hour decross-linking). n = 4 for each method. Standard deviations are shown.
In many cases, significant variability in DNA yields was observed among replicate samples purified using the same method. This is not surprising due to the inherent variability of the amount of tissue in any given FFPE tissue section. In general, increasing the decross-linking time for the ReliaPrep™ FFPE gDNA Miniprep System from 1 hour to 4 hours resulted in increased DNA recovery based on the amount of amplifiable DNA. While the amount of amplifiable DNA extracted with the ReliaPrep™ FFPE gDNA Miniprep System and a 1-hour decross-linking was typically lower than that with the QIAamp® DNA FFPE Tissue Kit, increasing the decross-linking time to 4 hours resulted in similar if not higher yields overall.
One of the most important parameters in qPCR assay design is amplicon size, as amplification of smaller fragments tends to be more efficient. Amplicon size is even more critical when the DNA to be amplified is purified from FFPE tissue. The DNA will likely be highly degraded, fragmented and potentially cross-linked, causing the amplification of larger amplicons to be even less efficient. We examined the effects of amplicon size when determining DNA yields from the various FFPE tissues using qPCR and GAPDH primer sets for 100bp and 300bp amplicons. The results are shown in Figure 3.
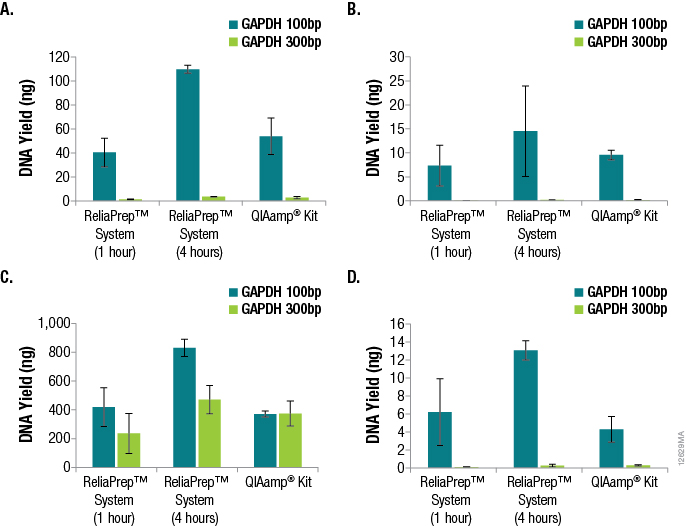
Figure 3. Effects of amplicon size when determining DNA yields from various FFPE tissues using qPCR and GAPDH primer sets for 100bp and 300bp amplicons. Panel A. DNA purified from lung tissue. Panel B. DNA purified from breast tissue. Panel C. DNA purified from colon tissue. Panel D. DNA purified from heart tissue. n = 4 for each method and primer set. Standard deviations are shown.
A significant decrease in DNA yield, as determined by qPCR, was observed when amplifying the 300bp GAPDH target compared to the 100bp GAPDH target, demonstrating the effect of amplicon size specifically when amplifying DNA from FFPE tissue samples. For the lung, breast and heart samples, a 15- to 100-fold difference in functional or amplifiable DNA was observed. The differences in yield based on amplicon sizes were less apparent for DNA from colon FFPE tissue (one- to twofold). It is likely that disparities in sample age, sample handling and formalin-fixation conditions attributed to these differences.
Methods for DNA purification from FFPE samples require a series of time-consuming preprocessing steps for deparaffinization, tissue lysis, decross-linking and RNase treatment. Many researchers prefer methods that are flexible, so identifying potential stopping points in the procedures can help researchers adjust to other time constraints when these methods can take hours to complete. We asked whether samples preprocessed using the ReliaPrep™ FFPE gDNA Miniprep System method can be stored overnight before purification with no loss of performance. To answer this question, we preprocessed samples (deparaffinization, tissue lysis and RNase treatment) using the standard protocol for the ReliaPrep™ FFPE gDNA Miniprep System, added BL Buffer and ethanol to the samples and stored the samples overnight at –20°C. The following day, the remaining purification steps (nucleic acid binding, column wash and elution) were completed. A comparison of the DNA recoveries between same-day processing and next-day processing is shown in Figure 4.
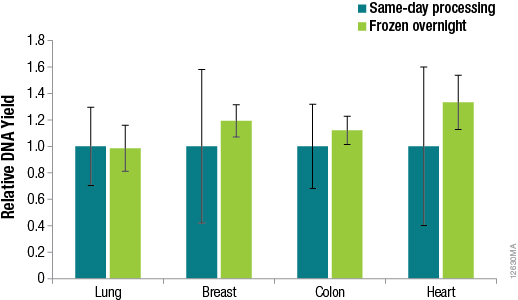
Figure 4. DNA yields from FFPE samples that were processed the same day or after overnight storage. DNA was quantified by qPCR using the GoTaq® qPCR Master Mix and human GAPDH primers for a 100bp amplicon and a human DNA standard. n = 4 for each method. Standard deviations are shown.
The results in Figure 4 confirm that no significant difference in DNA recoveries was observed between same-day processing and next-day processing following overnight storage of samples at –20°C. The ability to pause between sample preprocessing and DNA purification provides the flexibility to perform the protocol at times that are most convenient for the researcher.
Conclusions
quick and highly effective manner. Increasing the decross-linking time for the ReliaPrep™ FFPE gDNA Miniprep System from 1 hour to 4 hours resulted in increased DNA recovery based on the amount of amplifiable DNA. When amplifying DNA from FFPE tissues, the importance of primer design was demonstrated, as differences in yield were observed when amplicons of different sizes were amplified. In addition, no difference in DNA recoveries was observed between same-day processing and next-day processing following overnight storage of preprocessed samples, highlighting the flexibility of the streamlined protocol.
Successful purification of functional or amplifiable DNA from formalin-fixed, paraffin-embedded tissue can be a difficult process, as the DNA is often highly cross-linked, degraded and fragmented mainly due to formalin fixation during tissue preservation. The ReliaPrep™ FFPE gDNA Miniprep System has many unique features that can overcome these challenges in a safe,
References
- ReliaPrep™ FFPE gDNA Miniprep System Technical Manual TM352, Promega Corporation.
- QIAamp® DNA FFPE Tissue Handbook, QIAGEN Corporation.