GeneMarker® HID Software Review
Richland County Sheriff’s Department, Columbia, South Carolina
Publication Date: 2015
Introduction
The use of a new software tool for the analysis of short tandem repeats (STR) for human identification other than an Applied Biosystems product is at first intimidating. I have personal experience with GeneScan®, Genotyper®, GeneMapper® ID and GeneMapper® ID-X software (and TrueAllele®, CyberGenetics) and have relied on these software packages since 1999. These platforms have evolved from two separate programs dealing with the raw and analyzed data separately on an Apple® computer to integrated software packages residing on servers running in the Microsoft Windows® environment.
The Richland County Sheriff’s Department’s (RCSD) DNA laboratory is constantly looking for the most efficient and cost-effective means to complete approximately 1,500 case requests per year. In 2012, the RCSD DNA laboratory completed an average of 128 cases per month. RCSD validated the PowerPlex® Fusion System (Promega) in the summer of 2013. The sensitivity and amount of information developed (24 versus 16 loci) increased, causing our efficiency to decrease. Hence in 2014, an average of 98 cases per month were completed. This decrease in efficiency was directly related to the increase in information gained from the new 24-plex kit and the increased sensitivity of the chemistry. With this information gain in a shorter analysis time, the bottleneck for forensic DNA laboratories continues to be data analysis and interpretation (in addition to other non-laboratory tasks). With current staffing and ever-shrinking budgets, any tool the forensic scientist can employ to increase efficiency is beneficial; hence, the Richland County Sheriff’s Department has acquired, validated and performance checked GeneMarker® HID software v2.7.1 (Softgenetics).
Why Invest the Time?
After the acquisition of an IntegenX RapidHIT® instrument in December of 2014, RCSD was introduced to GeneMarker® HID analysis software. GeneMarker® HID comes bundled with this instrument and allowed the RCSD DNA laboratory to familiarize the staff with some of the features of this software platform. Several factors were considered for the purchase of GeneMarker® HID software. First, GeneMarker® HID is NDIS approved as an expert system by the Federal Bureau of Investigations (FBI). Second, the cost of the software package was approximately one third the costs to deploy for the RCSD versus other software platforms. Third, implementation did not require any infrastructure upgrades at the RCSD. Workstations running Windows® 7 can run the software without upgrade or modification. GeneMarker® HID can import files (.fsa and .hid) from common genetic analyzer sources and analyze them using many familiar parameters. Fourth, SoftGenetics provided online training through GoToMeeting™, offering customized training time slots to accommodate the laboratory personnel’s schedules. In addition, the support staff at SoftGenetics answered many additional questions and “how-tos” via e-mail. Finally, before any side-by-side comparisons or performance checks were conducted, the initial perception was that the GeneMarker® HID software was faster and more efficient at handling the substantial data produced by RapidHIT® GlobalFiler® Express Kit and PowerPlex® Fusion Systems.
Perception is Reality and Process Improvement
The following functions of GeneMarker® HID have not been thoroughly or empirically studied due to time and manpower constraints in our case-working laboratory but rather represent an overall impression or perception of how the software functions in the RCSD DNA laboratory’s process. GeneMarker® HID functions similarly to a “thin” client. To the end-user this means that project files can be easily saved on network locations or moved via thumb/USB-drive from computer to computer and opened easily via a simple double-click. All raw data and run parameters are contained in the project file. Technical reviewers can open these projects for review on different workstations, and all information and raw data are contained in the project file. There is no need to open the software program, search (or import) a project for analysis or review, and then begin the analysis or review. These differences in opening, analyzing and processing seem insignificant; however, all add up quickly in the data crunch that a DNA analyst endures day to day.
The RCSD DNA laboratory amplifies and processes items from several different cases at one time in a batch. After amplification and electrophoresis, all data from a run are analyzed, verified and technically reviewed. This process is facilitated by several features found in GeneMarker® HID. Hence, all samples can be printed (new page for each sample) with one click. In addition, printing “templates” can be created, ensuring different features for different samples, applications or kits. For example, different templates for the PowerPlex® Fusion System and RapidHIT® GlobalFiler® Express Kit can be created since one of these kits is 5-dye and one is 6-dye. Or a different template for printing the ladder samples can be created with specific formatting. The print templates can save specific parameters for printing to facilitate consistent printouts for varied electropherograms.
One of the first noticeable features of GeneMarker® HID was its ability to handle baselines and pull-up. The baseline subtraction function removes the baseline completely so that the Y axis will be raised above the noise level. It uses 20% of lowest intensities (to the right of the beginning of the range) and looks at the trace in 500- to 600-frame sections (1) . The pull-up correction feature removes peaks caused by bleedthrough to other wavelengths (1) . Figure 1 shows one example of the effectiveness of these functions. The blue dye from a single-source sample amplified with the PowerPlex® Fusion System and analyzed with GeneMapper® ID v3.2.1 is on the top panel, and the same sample analyzed with GeneMarker® HID v2.7.1 is on the bottom panel. GeneMapper® ID v3.2.1 was set up with an analytical threshold of 75RFU, light smoothing, baseline window of 31, minimum peak half width 2pt, polynomial degree of 3 and a peak size window of 13pt. GeneMarker® HID was set up with an analytical threshold of 75RFU, smoothing, peak saturation, baseline subtraction, pull-up correction and spike removal. Both used Local Southern size calling and identical stutter filters for the given loci. The practical result of these functions is less analyst input, and therefore greater efficiency, because analysts do not have to address (by documenting or deselecting) the sample artifacts.
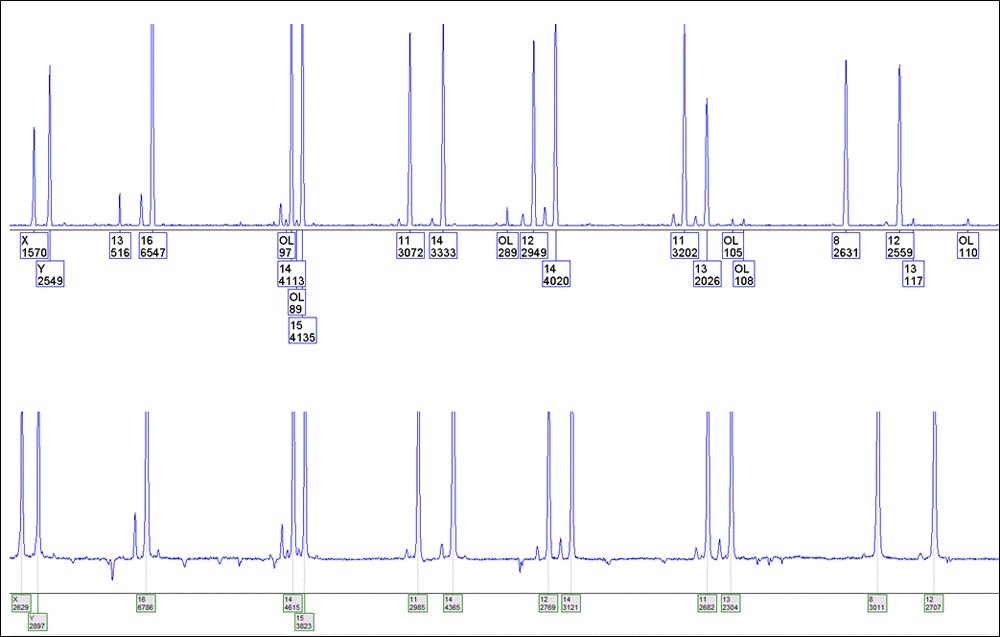 Figure 1. The blue dye from a single-source sample amplified with the PowerPlex® Fusion System and analyzed with GeneMapper® ID v3.2.1 (top panel) and GeneMarker® HID v2.7.1 (bottom panel).
Figure 1. The blue dye from a single-source sample amplified with the PowerPlex® Fusion System and analyzed with GeneMapper® ID v3.2.1 (top panel) and GeneMarker® HID v2.7.1 (bottom panel). Other convenience features include, but are not necessarily unique to GeneMarker® HID platform, identification of ladders, positive controls and negatives, and the ability to segregate these samples from the casework sample files. File name grouping and recent project memory are also useful. The “recent” project memory functions similarly to other Microsoft Office® programs. When opening the program from the Windows® Start button, an arrow will appear to the right of the GeneMarker® HID icon, indicating “recent” projects. Hovering over the arrow will reveal these projects for selection and one-click opening (Figure 2). GeneMarker® HID automatically adjusts the Y axis to the highest peak within the electropherogram view. This function suitably formats the electropherograms for printing and inclusion in case documentation, eliminating the need to manually scale each color with the mouse zoom for each sample. Another unique function allows the user to display (and print) the peak height ratios on the electropherograms for heterozygous loci (Figure 3). Users can set quality flags to indicate when peak height ratios are below the laboratory’s acceptable limits (one of several quality-flagging parameters).
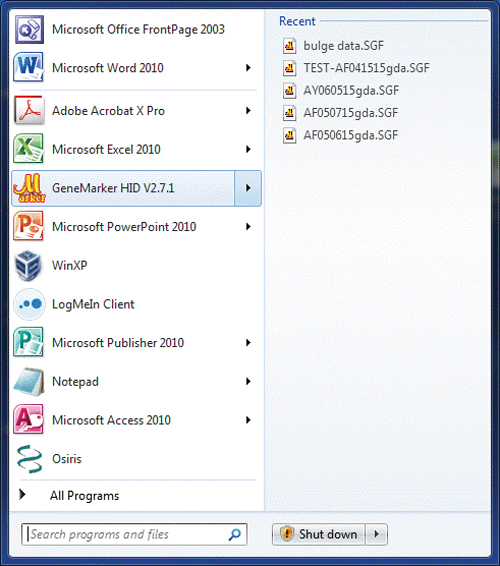 Figure 2. Opening recent GeneMarker® HID projects.
Figure 2. Opening recent GeneMarker® HID projects.
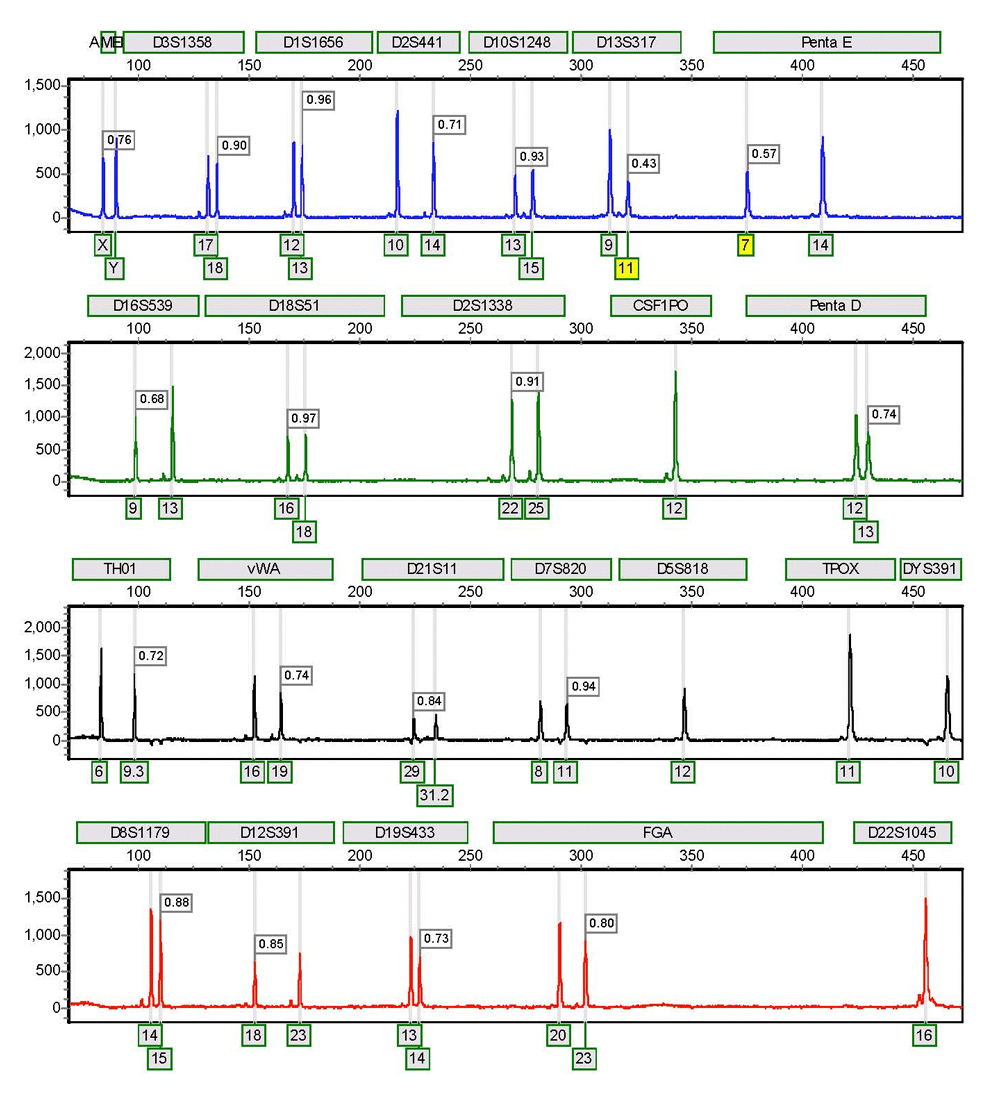 Figure 3. Peak height ratio labeling of heterozygous peaks in GeneMarker® HID software.
Figure 3. Peak height ratio labeling of heterozygous peaks in GeneMarker® HID software. Yellow flags indicate peak height ratios below 60%.
GeneMarker® HID also includes a function that exports common message format (CMF) files (versions 3.2., 3.0 and 1.0) to CODIS. This tool accommodates less-than-full STR profiles. Some CODIS export tools create a CMF file that contains null information at loci without alleles, causing their subsequent import into CODIS to crash. This feature is important for the PowerPlex® Fusion System (and GlobalFiler® PCR Amplification Kit) due to the fact that these kits contain male-specific loci [DYS391 and YIndel (GlobalFiler® Kit)]. Since females will not have alleles at these male-specific loci, CMF file creation must account for this scenario and create a properly formatted file for CODIS import.
In addition to the features discussed above, GeneMarker® HID contains some “advanced” features that require additional training and will not be discussed in this review (i.e., Paternity tool, Contamination tool, Mixture analysis tool, Project comparison tool et al.).
Conclusion
In conclusion, in the never-ending search to increase efficiency, decrease analyst input and fatigue, and decrease cost, the RCSD has employed a new software tool. GeneMarker® HID is a robust alternative to the established software platforms for the analysis of STRs for human identification in forensic DNA laboratories. Employing this software in combination with the PowerPlex® Fusion chemistry results in greater efficiency by decreasing analysis time despite the increase in information gained using the new 24-plex kits [expanded CODIS core loci required in 2017 (2) ]. The software features can be utilized to create a more efficient process and lead to a decrease in interpretation and review time for the analyst.
Acknowledgments
The author would like to thank Teresa Snyder-Leiby of SoftGenetics for technical support.
Article References
- GeneMarker® HID STR Human Identity Software User Manual, Softgenetics.
- Federal Bureau of Investigation (2015) Planned process and timeline for implementation of additional CODIS core loci.
How to Cite This Article
Scientific Style and Format, 7th edition, 2006
Amick, G.D. GeneMarker® HID Software Review. [Internet] 2015. [cited: year, month, date]. Available from: https://www.promega.com/resources/profiles-in-dna/2015/genemarker-hid-software-review/
American Medical Association, Manual of Style, 10th edition, 2007
Amick, G.D. GeneMarker® HID Software Review. Promega Corporation Web site. https://www.promega.com/resources/profiles-in-dna/2015/genemarker-hid-software-review/ Updated 2015. Accessed Month Day, Year.
Contribution of an article to Profiles in DNA does not constitute an endorsement of Promega products.
PowerPlex is a registered trademark of Promega Corporation.
Apple is a registered trademark of Apple Inc., registered in the U.S. and other countries. GeneMapper, GeneScan and Genotyper are registered trademarks of Applied Biosystems. GeneMarker is a registered trademark of SoftGenetics. GlobalFiler is a registered trademark of Life Technologies Corporation. Office and Windows are registered trademarks of Microsoft Corporation. RapidHIT is a registered trademark of IntegenX Inc. TrueAllele is a registered trademark of Cybergenetics, Inc.
Products may be covered by pending or issued patents or may have certain limitations. More information.
All prices and specifications are subject to change without prior notice.
Product claims are subject to change. Please contact Promega Technical Services or access the Promega online catalog for the most up-to-date information on Promega products.