High-Throughput Genetic Data Gathering From Bone Samples Recovered from an Air Crash Site and Processed Using a Semi-Automated DNA Purification Platform
1Servicio de Huellas Digitales Geneticas and Department of Genetics and Molecular Biology, School of Pharmacy and Biochemistry, Universidad de Buenos Aires, Argentina, 2Cuerpo Médico Forense de la Justicia Nacional, Department of Justice, Argentina.
Publication Date: 2013
Abstract
DNA-based analysis constitutes the key approach in the challenging task of identifying human remains from mass fatalities. In the present case, we introduced a workflow that allowed effective and fast identification of the complete set of highly fragmented human remains (N = 418) from an air crash in Río Negro Province, Argentina, on May 18, 2011, in which all 19 passengers and 3 crew members were killed. This process included DNA extraction from bone using a semi-automated DNA purification system, DNA quantitation by real-time PCR using commercial kits and data analysis using expert software. The implementation of this strategy allowed us to achieve our objective in a very short time.
Introduction
DNA-assisted identification is the gold standard for personal identification in any forensic scenario. Mass fatalities where only highly fragmentary remains are recovered at the disaster site represent one of the most challenging analytical situations. These cases require coordinated sampling in contrasting landscapes: Collection of samples at the disaster site, which can demand considerable time depending on the case, versus collection of reference samples donated by putative family members seeking to identify their missing relatives. This approach has been successfully applied in various situations in the last 20 years, such as victim identification in terrorist attacks (1) (2) (3) , the World Trade Center attack on September 11, 2001 (4) (5) , air crashes (6) , natural disasters (7) (8) and mass graves (9) . Optimization of the methodological platforms and improvements in sensitivity reduced the time required for complete identification. In this paper, we describe the methodological and interpretative approaches employed for identifying a set of more than 400 highly fragmented human remains, mostly represented by small bone fragments, belonging to the 22 fatalities.
Case description
On May 18, 2011, a SAAB 340 airplane belonging to Sol Airlines exploded at high altitude of more than 2,000 meters near Los Menucos (40°60´S, 68°07´W), in Río Negro province, Argentina. All 19 passengers and 3 crew members were killed. The remains were distributed throughout an area of about 1,000 square meters. Retrieval of all fragmentary remains took place during a considerable span of time, ending nine months after the accident.
Material and Methods
Human remains: Highly burned and fragmented remains were received at the Buenos Aires Forensic Mortuary. DNA laboratory members participated in selecting all of the samples to be used for DNA typing. Only one body was found almost completely intact, while the rest were severely fragmented. No complete skulls were found, but a high number of cranial bones were retrieved, indicating that an intense explosion had occurred, resulting in body and cranial burst. The human remains from the disaster area were sent for analysis on five different dates. The first two sets of samples included primarily soft tissues, whereas the remaining samples were mostly bone fragments. A total of 418 samples were selected for DNA typing using the following criteria: a) samples had to be anatomically recognizable, b) samples composed of bone, muscle and skin were considered a single part provided they were linked by tissue bridges and c) priority was given to muscle during the first months of testing, then later to bone samples. Reception dates and sample types are summarized in Figure 1.
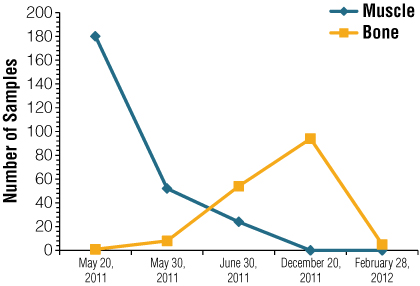 Figure 1. Summary of sample date reception and number of samples submitted to the Buenos Aires Forensic Mortuary for analysis.
Figure 1. Summary of sample date reception and number of samples submitted to the Buenos Aires Forensic Mortuary for analysis. At the forensic mortuary, all cadaver samples were carefully washed with tap water, numbered and photographed, and small tissue fragments were taken and placed in 50ml tubes containing a volume of solid table salt equivalent to 25ml (10) before being sent to the DNA laboratory. The remaining material was kept at –20°C for preservation in labeled heat-sealed plastic bags.
Family members: Members of the 22 families provided antemortem information such as radiographs, dental records, tattoos and descriptions of personal belonging. A total of 45 reference blood samples were obtained and spotted onto Whatman 3MM filter paper. All donors signed an informed consent statement approved by the Bioethics Committee of the Universidad de Buenos Aires School of Pharmacy and Biochemistry, Argentina. Although all relatives were sampled, first-degree relatives were given priority.
DNA extraction: All samples were extracted using the DNA IQ™ System and the semi-automated Maxwell® 16 Instrument (Promega Corporation). DNA from either muscle or bone was extracted from material that had been rinsed with distilled water to remove residual solid salt. Bone fragments were cut with a circular saw and muscles cut with a scalpel. Before adding the extraction buffer, tissues were weighed and adjusted to 100mg to standardize the amount of extracted DNA and reduce the need for real-time PCR quantitation. Tissue was placed in 2ml microcentrifuge tubes with 500µl of extraction buffer: 0.5% TEC-SDS (10mM Tris buffer [pH 7.6], 1mM ethylenediamine tetraacetic acid disodium salt [pH 8.0], 100mM NaCl and 0.5% w/v sodium dodecyl sulfate) and 10µl (20mg/ml) of Proteinase K (Promega Corporation). Samples were incubated overnight at 56°C and agitated at 750rpm in a Vortemp 1550 (LabLine, USA). A volume of 250µl of lysis buffer with 1M DTT (dithiothreitol, Promega Corporation) was added to each sample following the manufacturer´s protocol. Samples were vortexed at maximum speed for 10 seconds. Finally, samples were placed into the Maxwell® 16 cartridges, and 30µl of elution buffer was added for sample recovery. Purified DNA samples were kept at 4°C.
Editor's Note: For optimal results, Promega recommends an elution volume of 50µl with the Maxwell® 16 Instrument.
DNA Quantitation: To reduce analysis time, DNA extraction from muscle tissues was standardized after the quantification of 50 random samples by real-time PCR using the Plexor® HY System (Promega Corporation) and a Corbett RotorGene 6000 (Corbett Life Science). We found that samples weighing 100mg were the most appropriate for DNA extraction.
DNA samples obtained from the bones recovered during the last eight months after the accident were quantitated because bone samples had lower DNA yields than undecomposed muscle.
DNA Typing: All samples were typed for autosomal markers using the PowerPlex® 16 HS System (Promega Corporation) following the manufacturer´s protocol. After analysis, aliquots of all samples with identical genotypes were pooled and re-amplified for confirmation of the identity of samples in each group. DNA in each pooled sample was quantified and adjusted to the optimal concentration. In cases where the Amelogenin results indicated a male genotype, pooled samples were haplotyped with the Y-Filer™ kit (Life Technologies/Applied Biosystems).
Capillary Electrophoresis: Fragment analysis was performed with an ABI PRISM® 3100-Avant Genetic Analyzer. Profile interpretation and genetic identity searches were carried out with the GeneMapper® ID-X expert software (Life Technologies/Applied Biosystems). This software allows quality evaluation of genetic profiles, detection of potential lab reference sample contamination and sample comparison to establish identity among samples and kinship with reference samples.
Results and Discussion
Analysis of the first 181 samples led to identification of the 22 victims by comparison with reference samples within a week. Additional remains were retrieved in the crash area during a period of over nine months and were sent for further analysis (Figure 1).
With the exception of the mechanical procedure for cutting fragments of bone or muscle tissue, the protocols employed for DNA extraction were identical for both material, including modifications to the incubation step.
DNA was extracted from bone without demineralization because we targeted the trabecular bone-derived cells in samples that were retrieved during a relatively short time period of up to nine months following the crash. Additionally, the accident area is a semi-desert region with very low temperatures during winter (May to September) and is very dry in the spring and summer when the bone samples were found.
DNA quantitation represents an important step when bone samples are analyzed. We highly recommend a real-time PCR-based approach for this purpose, especially a highly standardized commercial kit. This step allows us to detect male material and potential inhibitors and adjust the amount of DNA to obtain the best profiles, reducing the chance of stochastic effects and avoiding split peaks.
No contamination was detected in any of the samples analyzed, as determined by the expert software. This software allowed us to group the 418 samples into 22 sample sets corresponding to the 22 victims and establish kinship relationships with the reference samples.
Bone DNA extraction has been improved since the beginning of the forensic DNA-based identification era (11) . Since then, a huge effort has been made to increase sensitivity, reduce inhibitory effects of environmental compounds and generate complete profiles from challenging samples.
In this paper we introduced some modifications that reduced processing time, intersample contamination and interpretation time. The strategy developed (Figure 2) proved to be optimal for a considerable number of samples from a reduced number of victims.
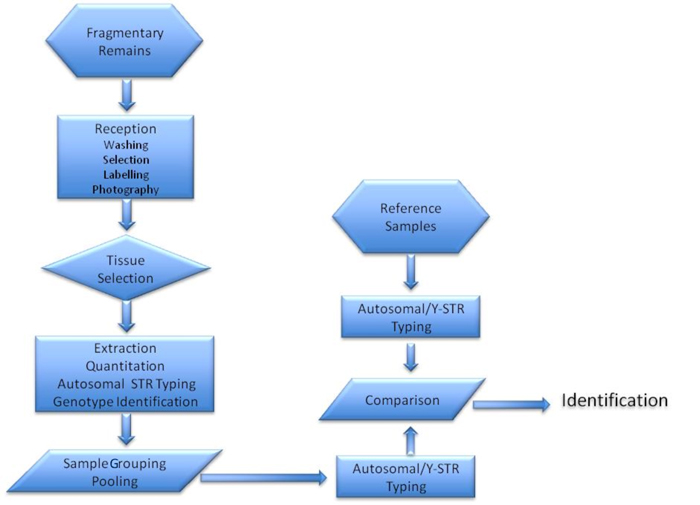 Figure 2. Flow chart describing the victim identification process.
Figure 2. Flow chart describing the victim identification process. Acknowledgements
DC, CB, MC and AS are members of the Career of Scientific Research CONICET; EA is a PhD post-graduate fellow at CONICET (National Research Council, Argentina).
Article References
- Corach, D. et al. (1995) Mass disasters: Rapid molecular screening of human remains by means of short tandem repeats typing. Electrophoresis 16, 1617–23.
- Corach, D. et al. (1997) Additional approaches to DNA typing of skeletal remains: The search for "missing" persons killed during the last dictatorship in Argentina. Electrophoresis 18, 1608–12.
- Ballantyne, J. (1997) Mass disaster genetics. Nat. Genet. 15, 329–31.
- Brenner, C.H. and Weir, B.S. (2003) Issues and strategies in the DNA identification of World Trade Center victims. Theor. Popul. Biol. 63, 173–8.
- Biesecker, L.G. et al. (2005) Epidemiology. DNA identifications after the 9/11 World Trade Center attack. Science 310, 1122–3.
- Hsu, C.M. et al. (1999) Identification of victims of the 1998 Taoyuan Airbus crash accident using DNA analysis. Int. J. Legal Med. 113, 43–6.
- Alonso, A. et al. (2005) Challenges of DNA profiling in mass disaster investigations. Croat. Med. J. 46, 540–8.
- Budowle, B., Bieber, F.R. and Eisenberg, A.J. (2005) Forensic aspects of mass disasters: Strategic considerations for DNA-based human identification. Legal Med. (Tokyo) 7, 230–43.
- Davoren, J. et al. (2007) Highly effective DNA extraction method for nuclear short tandem repeat testing of skeletal remains from mass graves. Croat. Med. J. 48, 478–85.
- Caputo, M., Bosio, L.A. and Corach, D. (2011) Long-term room temperature preservation of corpse soft tissue: An approach for tissue sample storage. Investig. Genet. 2, 17.
- Jeffreys, A.J., Wilson, V. and Thein, S.L. (1985) Hypervariable 'minisatellite' regions in human DNA. Nature 314, 67–73.
How to Cite This Article
Scientific Style and Format, 7th edition, 2006
Corach, D. et al. High-Throughput Genetic Data Gathering From Bone Samples Recovered from an Air Crash Site and Processed Using a Semi-Automated DNA Purification Platform. [Internet] 2013. [cited: year, month, date]. Available from: https://www.promega.com/resources/profiles-in-dna/2013/high-throughput-genetic-data-gathering-from-bone-samples-recovered-from-an-air-crash-site/
American Medical Association, Manual of Style, 10th edition, 2007
Corach, D. et al. High-Throughput Genetic Data Gathering From Bone Samples Recovered from an Air Crash Site and Processed Using a Semi-Automated DNA Purification Platform. Promega Corporation Web site. https://www.promega.com/resources/profiles-in-dna/2013/high-throughput-genetic-data-gathering-from-bone-samples-recovered-from-an-air-crash-site/ Updated 2013. Accessed Month Day, Year.
Contribution of an article to Profiles in DNA does not constitute an endorsement of Promega products.
Maxwell, Plexor and PowerPlex are registered trademarks of Promega Corporation. DNA IQ is a trademark of Promega Corporation.
ABI PRISM and GeneMapper are registered trademarks of Applied Biosystems.